













| General Information: |
|
| Name(s) found: |
GSPE_ECOLI
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli |
| Length: | 493 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRIHSPYPAS WALAQRIGYL YSEGEIIYLA DTPFERLLDI QRQVGQCQTM TSLSQADFEA 60
61 RLEAVFHQNT GESQQIAQDI DQSVDLLSLS EEMPANEDLL NEDSAAPVIR LINAILSEAI 120
121 KETASDIHIE TYEKTMSIRF RIDGVLRTIL QPNKKLAALL ISRIKVMARL DIAEKRIPQD 180
181 GRISLRIGRR NIDVRVSTLP SIYGERAVLR LLDKNSLQLS LNNLGMTAAD KQDLENLIQL 240
241 PHGIILVTGP TGSGKSTTLY AILSALNTPG RNILTVEDPV EYELEGIGQT QVNTRVDMSF 300
301 ARGLRAILRQ DPDVVMVGEI RDTETAQIAV QASLTGHLVL STLHTNSASG AVTRLRDMGV 360
361 ESFLLSSSLA GIIAQRLVRR LCPQCRQFTP VSPQQAQMFK YHQLAVTTIG TPVGCPHCHQ 420
421 SGYQGRMAIH EMMVVTPELR AAIHENVDEQ ALERLVRQQH KALIKNGLQK VISGDTSWDE 480
481 VMRVASATLE SEA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
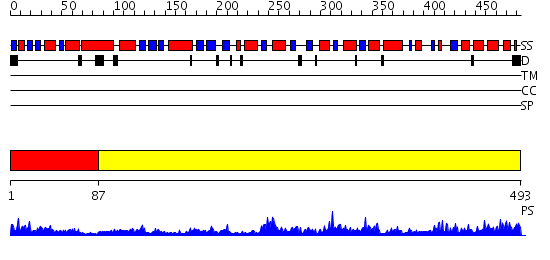
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..86] | 4.69897 | X-ray structure of the general secretion pathway complex of the N- terminal domain of EpsE and the cytosolic domain of EpsL of Vibrio cholerae | |
| 2 | View Details | [87..493] | 124.0 | Crystal Structure of Vibrio cholerae putative NTPase EpsE |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)