













| General Information: |
|
| Name(s) found: |
DPOLA_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1492 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
negative regulation of flower development
[NAS]
leaf morphogenesis [IMP] |
| Molecular Function: |
DNA-directed DNA polymerase activity
[ISS][TAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSGDNSTETG RRRSRGAEAS SRKDTLERLK AIRQGGIRSA SGGGYDIRLQ KPIFDTVDDE 60
61 EYDALVSRRR EEARGFVVED GEGGDLGYLD EGEEEDWSKP SGPESTDESD DGGRFSGRLK 120
121 KKKKGKEQTQ QPQVKKVNPA LKAAATITGE GRLSSMFTSS SFKKVKETDK AQYEGILDEI 180
181 IAQVTPDESD RKKHTRRKLP GTVPVTIFKN KKLFSVASSM GMKESEPTPS TYEGDSVSMD 240
241 NELMKEEDMK ESEVIPSETM ELLGSDIVKE DGSNKIRKTE VKSELGVKEV FTLNATIDMK 300
301 EKDSALSATA GWKEAMGKVG TENGALLGSS SEGKTEFDLD ADGSLRFFIL DAYEEAFGAS 360
361 MGTIYLFGKV KMGDTYKSCC VVVKNIQRCV YAIPNDSIFP SHELIMLEQE VKDSRLSPES 420
421 FRGKLHEMAS KLKNEIAQEL LQLNVSNFSM APVKRNYAFE RPDVPAGEQY VLKINYSFKD 480
481 RPLPEDLKGE SFSALLGSHT SALEHFILKR KIMGPCWLKI SSFSTCSPSE GVSWCKFEVT 540
541 VQSPKDITIL VSEEKVVHPP AVVTAINLKT IVNEKQNISE IVSASVLCFH NAKIDVPMPA 600
601 PERKRSGILS HFTVVRNPEG TGYPIGWKKE VSDRNSKNGC NVLSIENSER ALLNRLFLEL 660
661 NKLDSDILVG HNISGFDLDV LLQRAQACKV QSSMWSKIGR LKRSFMPKLK GNSNYGSGAT 720
721 PGLMSCIAGR LLCDTDLCSR DLLKEVSYSL TDLSKTQLNR DRKEIAPNDI PKMFQSSKTL 780
781 VELIECGETD AWLSMELMFH LSVLPLTLQL TNISGNLWGK TLQGARAQRI EYYLLHTFHS 840
841 KKFILPDKIS QRMKEIKSSK RRMDYAPEDR NVDELDADLT LENDPSKGSK TKKGPAYAGG 900
901 LVLEPKRGLY DKYVLLLDFN SLYPSIIQEY NICFTTIPRS EDGVPRLPSS QTPGILPKLM 960
961 EHLVSIRKSV KLKMKKETGL KYWELDIRQQ ALKLTANSMY GCLGFSNSRF YAKPLAELIT1020
1021 LQGRDILQRT VDLVQNHLNL EVIYGDTDSI MIHSGLDDIE EVKAIKSKVI QEVNKKYRCL1080
1081 KIDCDGIYKR MLLLRKKKYA AVKLQFKDGK PCEDIERKGV DMVRRDWSLL SKEIGDLCLS1140
1141 KILYGGSCED VVEAIHNELM KIKEEMRNGQ VALEKYVITK TLTKPPAAYP DSKSQPHVQV1200
1201 ALRMRQRGYK EGFNAKDTVP YIICYEQGNA SSASSAGIAE RARHPDEVKS EGSRWLVDID1260
1261 YYLAQQIHPV VSRLCAEIQG TSPERLAECL GLDPSKYRSK SNDATSSDPS TSLLFATSDE1320
1321 ESKKPATPET EESDSTFWLK LHCPKCQQED STGIISPAMI ANQVKRQIDG FVSMYYKGIM1380
1381 VCEDESCKHT TRSPNFRLLG ERERGTVCPN YPNCNGTLLR KYTEADLYKQ LSYFCHILDT1440
1441 QCSLEKMDVG VRIQVEKAMT KIRPAVKSAA AITRSSRDRC AYGWMQLTDI VI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
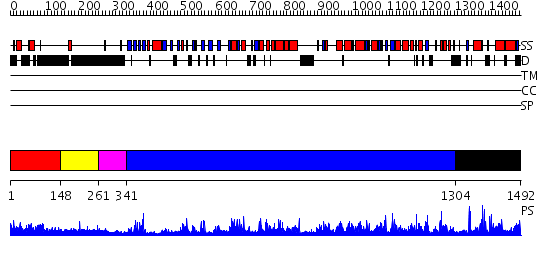
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..147] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [148..260] | 4.125996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [261..340] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [341..1303] | 149.0 | Exonuclease domain of family B (archaeal and phage) DNA polymerases; T4-like DNA polymerase | |
| 5 | View Details | [1304..1492] | 1.081987 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)