













| General Information: |
|
| Name(s) found: |
CLSY4_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1132 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: |
nucleic acid binding
[IEA]
DNA binding [IEA][ISS] ATP binding [IEA][ISS] helicase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDMTSCVARR TRSRTESYLN SILNKSKGIS GEEEDQSLGC VNSRTEKRRV NMRDACSPSP 60
61 RKKKRRRRKD DDDDVVFVRT EYPEGKRDDE NVGSTSGNLQ SKSFDFGDRV CDFDADDRNL 120
121 GCEEKASNFN PIDDDDDVVF VGTVQRENDH VEDDDNVGSA SVISPRVCDF DEDDAKVSGK 180
181 ENPLSPDDDD DVVFLGTIAG ENQHVEDVNA GSEVCDILLD DANLRGEEKT YVSDEVVSLS 240
241 SSSDDEEDPL EELGTDSREE VSGEDRDSGE SDMDEDANDS DSSDYVGESS DSSDVESSDS 300
301 DFVCSEDEEG GTRDDATCEK NPSEKVYHHK KSRTFRRKHN FDVINLLAKS MLESKDVFKE 360
361 DIFSWDKIAE VDSREDPVVR ESSSEKVNEH GKPRERRSFH RVREKNHLNG ESFYGGEKLC 420
421 DGEETINYST EDSPPLNLRF GCEEPVLIEK TEEEKELDSL WEDMNVALTL EGMHSSTPDK 480
481 NGDMLCSKGT HDFVLDDEIG LKCVHCAYVA VEIKDISPAM DKYRPSVNDN KKCSDRKGDP 540
541 LPNRLEFDAS DPSSFVAPLD NIEGTVWQYV PGIKDTLYPH QQEGFEFIWK NLAGTTKINE 600
601 LNSVGVKGSG GCIISHKAGT GKTRLTVVFL QSYLKRFPNS HPMVIAPATL MRTWEDEVRK 660
661 WNVNIPFYNM NSLQLSGYED AEAVSRLEGN RHHNSIRMVK LVSWWKQKSI LGISYPLYEK 720
721 LAANKNTEGM QVFRRMLVEL PGLLVLDEGH TPRNQSSLIW KVLTEVRTEK RIFLSGTLFQ 780
781 NNFKELSNVL CLARPADKDT ISSRIHELSK CSQEGEHGRV NEENRIVDLK AMIAHFVHVH 840
841 EGTILQESLP GLRDCVVVLN PPFQQKKILD RIDTSQNTFE FEHKLSAVSV HPSLYLCCNP 900
901 TKKEDLVIGP ATLGTLKRLR LKYEEGVKTK FLIDFIRISG TVKEKVLVYS QYIDTLKLIM 960
961 EQLIAECDWT EGEQILLMHG KVEQRDRQHM IDNFNKPDSG SKVLLASTKA CSEGISLVGA1020
1021 SRVVILDVVW NPSVESQAIS RAFRIGQKRA VFIYHLMVKD TSEWNKYCKQ SEKHRISELV1080
1081 FSSTNEKDKP INNEVVSKDR ILDEMVRHEK LKHIFEKILY HPKKSDMNTS FF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
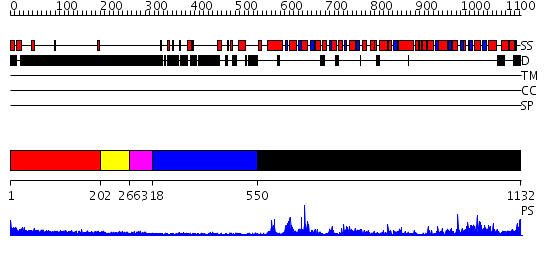
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..201] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [202..265] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [266..317] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [318..549] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [550..1132] | 90.39794 | Structure of the SWI2/SNF2 chromatin remodeling domain of eukaryotic Rad54 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)