













| General Information: |
|
| Name(s) found: |
osm-3 /
CE31568
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 699 amino acids |
Gene Ontology: |
|
| Cellular Component: |
perinuclear region of cytoplasm
[IDA neuronal cell body [IDA neuron projection [IDA microtubule associated complex [IEA microtubule [IDA |
| Biological Process: |
regulation of insulin receptor signaling pathway
[IGI microtubule-based movement [IEA |
| Molecular Function: |
ATPase activity
[ISS ATP binding [IEA microtubule motor activity [IEA protein binding [ISS plus-end-directed microtubule motor activity [ISS microtubule binding [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAESVRVAVR CRPFNQREKD LNTTLCVGMT PNVGQVNLNA PDGAAKDFTF DGAYFMDSTG 60
61 EQIYNDIVFP LVENVIEGYN GTVFAYGQTG SGKTFSMQGI ETIPAQRGVI PRAFDHIFTA 120
121 TATTENVKFL VHCSYLEIYN EEVRDLLGAD NKQKLEIKEQ PDRGVYVAGL SMHVCHDVPA 180
181 CKELMTRGFN NRHVGATLMN KDSSRSHSIF TVYVEGMTET GSIRMGKLNL VDLAGSERQS 240
241 KTGATGDRLK EATKINLSLS ALGNVISALV DGKSKHIPYR DSKLTRLLQD SLGGNTKTIM 300
301 IACVSPSSDN YDETLSTLRY ANRAKNIKNK PTINEDPKDA LLREYQEEIA RLKSMVQPGA 360
361 VGVGAPAQDA FSIEEERKKL REEFEEAMND LRGEYEREQT SKAELQKDLE SLRADYERAN 420
421 ANLDNLNPEE AAKKIQQLQD QFIGGEEAGN TQLKQKRMKQ LKEAETKTQK LAAALNVHKD 480
481 DPLLQVYSTT QEKLDAVTSQ LEKEVKKSKG YEREIEDLHG EFELDRLDYL DTIRKQDQQL 540
541 KLLMQIMDKI QPIIKKDTNY SNVDRIKKEA VWNEDESRWI LPEMSMSRTI LPLANNGYMQ 600
601 EPARQENTLL RSNFDDKLRE RLAKSDSENL ANSYFKPVKQ INVINKYKSD QKLSTSKSLF 660
661 PSKTPTFDGL VNGVVYTDAL YERAQSAKRP PRLASLNPK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
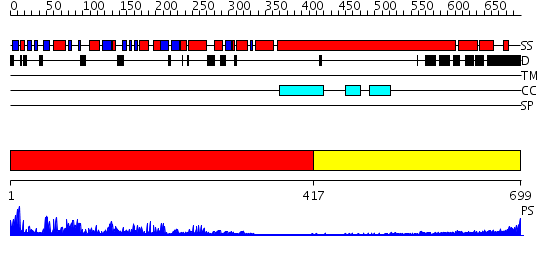
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..416] | 89.0 | Kinesin | |
| 2 | View Details | [417..699] | 8.154902 | Heavy meromyosin subfragment |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)