













| General Information: |
|
| Name(s) found: |
mut-2 /
CE11740
[WormBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 441 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
embryonic development ending in birth or egg hatching
[IMP meiotic chromosome segregation [IMP negative regulation of transposition, DNA-mediated [IMP RNA interference [IMP reproduction [IMP |
| Molecular Function: |
nucleotidyltransferase activity
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQPNKDQRS PEDIDFKVKA NPKAFHKFNG KFQRVLRDHE DDFNILSISM QDHFDTTKQP 60
61 KEEFGKKMDW CYQLKNIISK NNPTWLFNIV PTGSTVTGLA TKNSDLDVAI HIPQAARVLE 120
121 QEERGRNITD DERQASWREI QLEILQIVRL NLQNDEQINS RINWEHGIQL VQAQIQILKV 180
181 MTVDGIDCDI SVVMDRFLSS MHNSFLIRHL AHIDGRFAPL CAIVKQWAAS TKVKDPKDGG 240
241 FNSYALVLLV IHFLQCGTFP PILPNLQEIF KKDNFIAWDD KVYPSILNFG APLPKPLPRI 300
301 APNNAPLARL FIEFLYYYSM FNFKENYIGA RPVMVMDRRT SQNNMVRSST NKEVCIQDPF 360
361 DEHNPGRTVR TLNRIKDVMR STYQKFLPVE GSEFTFPTLD DIINMSPEVP KPSRAEVRNF 420
421 RAEATEVNRV GLGVYCRNSF V |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
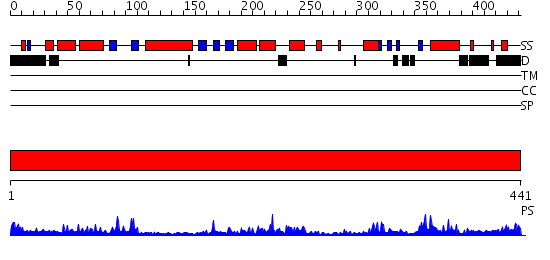
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..441] | 57.69897 | Poly(A) polymerase, middle domain; Poly(A) polymerase N-terminal, catalytic domain; Poly(A) polymerase, C-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)