













| General Information: |
|
| Name(s) found: |
SPCC1442.07c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 282 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA |
| Biological Process: |
protein sumoylation
[ISS cellular response to stress [IEP response to DNA damage stimulus [ISS |
| Molecular Function: |
zinc ion binding
[ISS metallopeptidase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MELKFSCRGN VIALSFNEND TVLDAKEKLG QEIDVSPSLI KLLYKGNLSD DSHLQDVVKN 60
61 ESKIMCLIRQ DKDIVNQAIS QLKVPDYSTN TYSLKPKKPH TTPKPASIYT FNELVVLDYP 120
121 HKDRALRYLE RLRDDTGIKK IMDSHRWTVP LLSEMDPAEH TRHDSKTLGL NHNQGAHIEL 180
181 RLRTDRYDGF RDYKTVKSTL IHELTHNVHG EHDSSFWELF RQLTKEADAA DLLGKPGSYV 240
241 SDRASYTPQQ DNDDEDQKNH RRDLLLAAAE RRKQSGSKVQ KE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
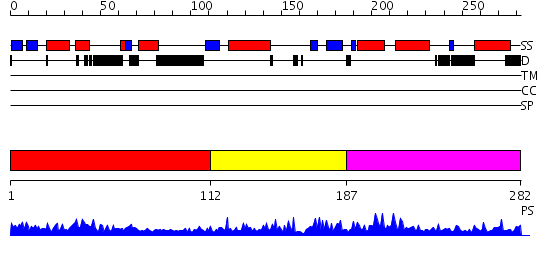
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..111] | 9.0 | Structure of the DNA repair protein hHR23a | |
| 2 | View Details | [112..186] | 5.657577 | No description for PF08325.1 was found. No confident structure predictions are available. | |
| 3 | View Details | [187..282] | 1.19 | No description for 2ckiA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)