













| General Information: |
|
| Name(s) found: |
slx8 /
SPBC3D6.11c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 269 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA SUMO-targeted ubiquitin ligase complex [IPI |
| Biological Process: |
DNA damage checkpoint
[IMP DNA recombination [IGI negative regulation of protein sumoylation [IMP protein ubiquitination [IDA |
| Molecular Function: |
ubiquitin-protein ligase activity
[IDA zinc ion binding [ISS protein binding [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPPAHKRDTN VRNLSAPYNI PSQSARVAAG NAAINRRRSS PVENSPGNGF PVSEDATDYP 60
61 SGTTSENESL PLNRAPRSLR EVASELAQEE TLPVETSDLN IDVESEVFDL EDINFQNDAD 120
121 DINQRFTYNN HPASVENSLT NVNSIHAQPT TISDMIDLTD ETSYDPRKQK FEQGKNPSTT 180
181 NAEIEKEEPS KKQVVPSSQR LADYKCVICL DSPENLSCTP CGHIFCNFCI LSALGTTAAT 240
241 QKCPVCRRKV HPNKVICLEM MLGSQKKKS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
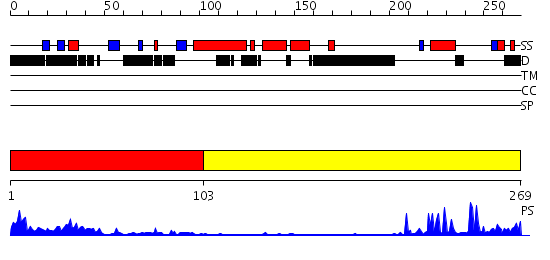
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..102] | 8.522879 | Structure of the SWI2/SNF2 chromatin remodeling domain of eukaryotic Rad54 | |
| 2 | View Details | [103..269] | 5.39794 | Crystal structure of the CHIP U-box E3 ubiquitin ligase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)