













| General Information: |
|
| Name(s) found: |
ade7 /
SPBC409.10
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 299 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [ISS cytosol [IDA |
| Biological Process: |
adenine biosynthetic process
[IMP 'de novo' IMP biosynthetic process [ISS |
| Molecular Function: |
ATP binding
[IEA]
phosphoribosylaminoimidazolesuccinocarboxamide synthase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALLKTSLDA PFTKIATGKV RDLYECVEFP DDLLFVATDR ISAYDFIMEN GIPEKGKVLT 60
61 KISEFWFDVL KPHVQTHLIT SRWEELPPVI TKHEELKDRS MLVKKYKILP IEAIVRGYIT 120
121 GSAWKEYQKQ GTVHGLNVPT GMKEAEAFPE PLFTPSTKAA EGHDENIHPD EVSKIVGPEL 180
181 AKQVAETSVK LYKIARDVAL KKGIIIADTK FEFGVDETTN KIVLVDEVLT PDSSRFWLAS 240
241 DYTVGKSPDS FDKQYLRNWL TANNLANKPN VSLPEDVVEA TRRKYVEAYE MITGKKWSY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
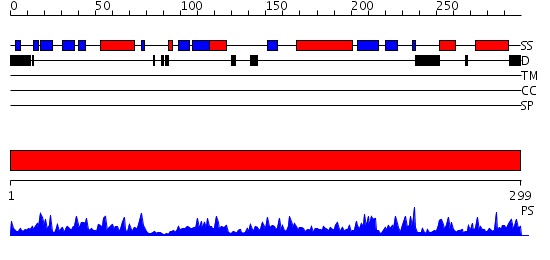
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..299] | 105.0 | SAICAR SYNTHASE |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)