













| General Information: |
|
| Name(s) found: |
SPAC56F8.03
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1079 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosolic small ribosomal subunit
[ISS cytosol [IDA |
| Biological Process: |
positive regulation of translational initiation
[IC translational initiation [ISS |
| Molecular Function: |
GTP binding
[IC translation initiation factor activity [ISS GTPase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGKKGKKSGY ADWEDDLGED ISGQNEYLDN TSQDSPQNDE LAEKSENLAV SSEKTTSKKK 60
61 KGKKNKGNKN QVSDDESQEL ESPQGPKELT AVTELDDDEF DYKPKKGKKG KKSKKVEEDD 120
121 EPQEIESPQG PKELTAVTEL DDDEFDYKPK KGKKGKKAQN NNESEAAAPP EIPEVRVKTK 180
181 KEKEREKKER EKLRKKQQQA KKKGSTGEDT LASSEVSSEV DISTPAENDS SAKGKQAAGS 240
241 KRKGPNVTAL QKMLEEKRAR EEEEQRIREE EARIAEEEKR LAEVEEARKE EARLKKKEKE 300
301 RKKKEEMKAQ GKYLSKKQKE QQALAQRRLQ QMLESGVRVA GLSNGEKKQK PVYTNKKKSN 360
361 RSGTSSISSS GILESSPATS ISVDEPQKDS KDDSEKVEKE TEVERKEENE AEAEAVFDDW 420
421 EAALEEPEVA ENNEVVTEKK ETDIKSDAVE HSIKDKEDSK TDKVDDIPQA APAESNVSES 480
481 DLRSPICCIL GHVDTGKTKL LDNLRRSNVQ EGEAGGITQQ IGATYFPIES IKQKTKVVNK 540
541 KGKLQYNIPG LLIIDTPGHE SFTNLRSRGT SLCNIAILVI DIMHGLEPQT IESIRLLRDQ 600
601 KTPFVVALNK VDRLYGWHSI KDNAIQDSLS KQKKAIQREF RDRVESIILQ LNEQGLNAAL 660
661 YFENKNLGRY VSLVPTSAQS GEGVPDLVAL LISLTQTRMS DRIKYITTLE CTVLEVKVIE 720
721 GLGATIDVIL SNGVLHEGDR IVLCGMGGPI ITTVRALLTP QPLKEMRVKS AYVHHKEIKA 780
781 AMGVKICAND LEKAVAGSRL LVVGPDDDEE DLAEEIMEDL ENLLGRIDTS GIGVSVQAST 840
841 LGSLEALLEF LKQMKIPVAS VNIGPVYKKD VMRCATMLEK AKEYALMLCF DVKVDRDAED 900
901 LAEQLGVKIF SANVIYHLFD AFTAHQKKIL EQKREESSDV AVFPCVLKTV AAFNKRDPII 960
961 LGVDVVEGVL RINTPIVAVK QLPNGEPQII ELGRVASLEM NHKPVDKVKK GQAGAGVAMK1020
1021 LESSGSQILF GRQVTESDAL YSHITRQSID SLKDPAFRDE VSRDEWQLII QLKKLFGII |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
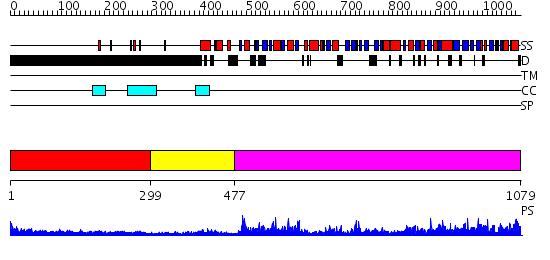
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..298] | 6.154902 | Heavy meromyosin subfragment | |
| 2 | View Details | [299..476] | 0.971 | crystal structure of the cytotoxic bacterial protein colicin B at 2.5 A resolution | |
| 3 | View Details | [477..1079] | 78.522879 | Initiation factor IF2/eIF5b, domains 2 and 4; Initiation factor IF2/eIF5b, domain 3; Initiation factor IF2/eIF5b, N-terminal (G) domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)