













| General Information: |
|
| Name(s) found: |
B-H2-PA /
FBpp0074203
[FlyBase]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 645 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA][NAS]
|
| Biological Process: |
sensory organ boundary specification
[IMP]
compound eye photoreceptor cell differentiation [IMP] bristle morphogenesis [IMP] eye-antennal disc morphogenesis [IMP] regulation of transcription, DNA-dependent [IEA][NAS] eye pigment granule organization [IMP] regulation of transcription [IEA] leg disc proximal/distal pattern formation [IMP] |
| Molecular Function: |
DNA binding
[NAS]
transcription factor activity [IEA][NAS] specific RNA polymerase II transcription factor activity [NAS] sequence-specific DNA binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTTMPPEMSA TTAAPVGSAP SATAHHPAAV GGGMPRPASP AVGSNTTTAT ATTATRSRFM 60
61 ITDILAGAAA ASAAAAAAAA ALAAASSGGG RGSPTDSERE QSLVAQHHHH HQQQQQHHHH 120
121 QQQQQQQQHQ QAALQQYIVQ QQQLLRFERE REREREREHY RERHSPPGNN PYAHHPMPPH 180
181 LLAHFPPAHY AVLQQQQQQQ QQQQHPHPHH LQLERERLEA LHRHGHGLTG DPAQHLSHLS 240
241 HLSHQQHHPH LHHPMHDERS RSPLMLQQLG GNGGNNNNNN NNSSSASNNN NNNNNSASAN 300
301 SNIISGNSSS SNNNNGSGNG NMLLGGPGSS ISGDQASTID DSDSDDCGGK DDDGDDSMKN 360
361 GSSANGDSSS HLSLSLSKKQ RKARTAFTDH QLQTLEKSFE RQKYLSVQDR MELANKLELS 420
421 DCQVKTWYQN RRTKWKRQTA VGLELLAEAG NYAAFQRLYG GATPYLSAWP YAAAAAAQSP 480
481 HGATPSAIDI YYRQAAAAAA MQKPSLPASY RMYQSSIPPG MSLPGMPAPP PPGAAPMLSG 540
541 YYAAAAAAAA SAGAQQQQQQ PPAASRSPAT SQSANSEADC ERTSSSSRQR LITPSPPLNP 600
601 GSPPHRERIN EEEDRERDEE RDIERERERE RERDEDDEEE LALEV |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
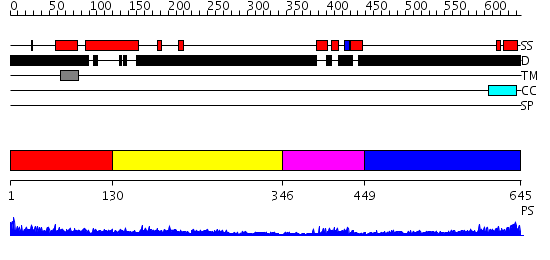
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..129] | 1.15 | The structure of a modular endo-beta-1,4-mannanase from Cellulomonas fimi explains the product specificity of glycoside hydrolase family 26 mannanases. | |
| 2 | View Details | [130..345] | 3.39794 | Sec24 | |
| 3 | View Details | [346..448] | 12.522879 | Homeobox protein hox-b1 | |
| 4 | View Details | [449..645] | 0.979 | Crystal Structure of Rat Synapsin I C Domain Complexed to Ca.ATP |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|