













| General Information: |
|
| Name(s) found: |
su(r)-PC /
FBpp0071273
[FlyBase]
|
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1031 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA]
|
| Biological Process: |
'de novo' pyrimidine base biosynthetic process
[IGI]
UMP biosynthetic process [IEA] oxidation reduction [IEA] |
| Molecular Function: |
electron carrier activity
[IEA]
dihydropyrimidine dehydrogenase (NADP+) activity [ISS][NAS] iron-sulfur cluster binding [IEA] dihydrouracil dehydrogenase (NAD+) activity [TAS] dihydroorotate oxidase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSPPQLLSKD SPDIEDLLSL NPRVKTQCSV VPTKQTKENK KHWKRNADHA APPCTTLAND 60
61 FSDIKHTTLS ERGALEEAAR CLKCADAPCQ KSCPTQLDIK SFITSIANKN FYGAAKAIFS 120
121 DNPLGLTCGM VCPTSDLCVG GCNLQASEAG PINIGGLQQF ATEVFKKMGV RQRRTPQAEA 180
181 NPLSQKIALV GGGPASLSCA TFLARLGYRD VTIYERRSYL GGLSAAEIPQ YRLPIDAVNF 240
241 EIDLVRDLGV RIETGRSLGT KDLTIQGLLS TGHDAVFVGI GLPEPKLNPI FAGLQPSNGF 300
301 YTSKNFLPLV SDGSKPGLCA CKAAAGLPKL HGNVIVLGAG DTAFDCATSA LRCGARRVFV 360
361 VFRKGSSGIR AVPEEVELAR DERCELLPYL SPRKVIVKDG LITAMEFCRT EQNENDEWVE 420
421 DEEQTQRLKA NFVISAFGSG LEDQDVKAAL APLQFRGELP VVDRVTMQSS VKQVFLGGDL 480
481 AGVANTTVES VNDGKVAAWS IHCQLQGLPL DTPAALPLFY TDIDAVDISV EMCGIRFENP 540
541 FGLASAPPTT STAMIRRAFE QGWGFVVTKT FGLDKDLVTN VSPRIVRGTT SGYKYGPQQG 600
601 CFLNIELISE KRAEYWLKSI GELKRDFPEK IVIASIMCSF NEEDWTELAI KAEQSGADAL 660
661 ELNLSCPHGM GERGMGLACG QDPELVEQIS RWVRKAVKLP FFIKLTPNIT DIVSIAAAAK 720
721 RGGADGGSAI NTVQGLMGLK ADSTAWPAIG KEQRTTYGGV SGNATRPMAL KAISDIANRV 780
781 PGFPILGIGG IDSGEVALQF IHAGATVLQI CSSVQNQDFT VIEDYCTALK ALLYLKANPP 840
841 PVDGPFWDGQ SPPTPVHQKG KPVVRLTGEG KATLGFFGPY QRQRDIKMAE LRSQKGALWD 900
901 AEQVKATPPA SNGAPNPAPR IKDVIGAALD KIGSYNKLDN KQQKVALIDD DMCINCGKCY 960
961 MTCADSGYQA IEFDKDTHIP HVNDDCTGCT LCVSVCPIID CITMVPKKIP HVIKRGVEEK1020
1021 IFYTHALSQC Q |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
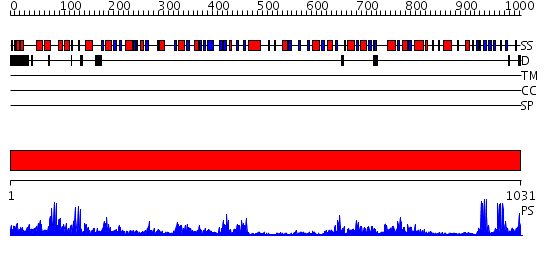
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..1031] | 1000.0 | Dihydropyrimidine dehydrogenase, N-terminal domain; Dihydropyrimidine dehydrogenase, domain 4; Dihydropyrimidine dehydrogenase, domain 3; Dihydropyrimidine dehydrogenase, domain 2; Dihydropyrimidine dehydrogenase, C-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)