













| General Information: |
|
| Name(s) found: |
mus308-PA /
FBpp0082131
[FlyBase]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 2059 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
DNA repair
[IEA][NAS][TAS]
DNA replication [IEA] |
| Molecular Function: |
DNA binding
[IEA]
ATP binding [IEA] DNA-directed DNA polymerase activity [ISS][NAS] ATP-dependent helicase activity [IEA] helicase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAFSQSFNFG NSTLMALEKG MQADDKENAQ PGNGNIQVQS AGNEVNSEIQ EINSEFFRDE 60
61 FSYEVNQAHK PAEQSVVNVS QVQQHMAVVS NQDSEDQSRS SALNDQICTQ SSFEGEDAGA 120
121 DAVLDQPNLD ENSFLCPAQD EEASEQLKED ILHSHSVLAK QEFYQEISQV TQNLSSMSPN 180
181 QLRVSPNSSR IREAMPERPA MPLDLNTLRS ISAWNLPMSI QAEYKKKGVV DMFDWQVECL 240
241 SKPRLLFEHC NLVYSAPTSA GKTLVSEILM LKTVLERGKK VLLILPFISV VREKMFYMQD 300
301 LLTPAGYRVE GFYGGYTPPG GFESLHVAIC TIEKANSIVN KLMEQGKLET IGMVVVDEVH 360
361 LISDKGRGYI LELLLAKILY MSRRNGLQIQ VITMSATLEN VQLLQSWLDA ELYITNYRPV 420
421 ALKEMIKVGT VIYDHRLKLV RDVAKQKVLL KGLENDSDDV ALLCIETLLE GCSVIVFCPS 480
481 KDWCENLAVQ LATAIHVQIK SETVLGQRLR TNLNPRAIAE VKQQLRDIPT GLDGVMSKAI 540
541 TYACAFHHAG LTTEERDIIE ASFKAGALKV LVATSTLSSG VNLPARRVLI RSPLFGGKQM 600
601 SSLTYRQMIG RAGRMGKDTL GESILICNEI NARMGRDLVV SELQPITSCL DMDGSTHLKR 660
661 ALLEVISSGV ANTKEDIDFF VNCTLLSAQK AFHAKEKPPD EESDANYIND ALDFLVEYEF 720
721 VRLQRNEERE TAVYVATRLG AACLASSMPP TDGLILFAEL QKSRRSFVLE SELHAVYLVT 780
781 PYSVCYQLQD IDWLLYVHMW EKLSSPMKKV GELVGVRDAF LYKALRGQTK LDYKQMQIHK 840
841 RFYIALALEE LVNETPINVV VHKYKCHRGM LQSLQQMAST FAGIVTAFCN SLQWSTLALI 900
901 VSQFKDRLFF GIHRDLIDLM RIPDLSQKRA RALFDAGITS LVELAGADPV ELEKVLYNSI 960
961 SFDSAKQHDH ENADEAAKRN VVRNFYITGK AGMTVSEAAK LLIGEARQFV QHEIGLGTIK1020
1021 WTQTQAGVEI ASRAIHDGGE VDLHMSLEEE QPPVKRKLSI EENGTANSQK NPRLETVVDT1080
1081 QRGYKVDKNI ANQSKMNPNL KEIDAQNKAR RNSTAHMDNL NPISNDPCQN NVNVKTAQPI1140
1141 ISNLNDIQKQ GSQIEKMKIN PATVVCSPQL ANEEKPSTSQ SARRKLVNEG MAERRRVALM1200
1201 KIQQRTQKEN QSKDQPIQAS RSNQLSSPVN RTPANRWTQS ENPNNEMNNS QLPRRNPRNQ1260
1261 SPVPNANRTA SRKVSNAEED LFMADDSFML NTGLAAALTA AESKIASCTE ADVIPSSQPK1320
1321 EPEVIGALTP HASRLKRSDQ LRSQRIQSPS PTPQREIEID LESKNESNGV SSMEISDMSM1380
1381 ENPLMKNPLH LNASHIMSCS KVDETASSFS SIDIIDVCGH RNAFQAAIIE INNATRLGFS1440
1441 VGLQAQAGKQ KPLIGSNLLI NQVAAAENRE AAARERVLFQ VDDTNFISGV SFCLADNVAY1500
1501 YWNMQIDERA AYQGVPTPLK VQELCNLMAR KDLTLVMHDG KEQLKMLRKA IPQLKRISAK1560
1561 LEDAKVANWL LQPDKTVNFL NMCQTFAPEC TGLANLCGSG RGYSSYGLDT SSAILPRIRT1620
1621 AIESCVTLHI LQGQTENLSR IGNGDLLKFF HDIEMPIQLT LCQMELVGFP AQKQRLQQLY1680
1681 QRMVAVMKKV ETKIYEQHGS RFNLGSSQAV AKVLGLHRKA KGRVTTSRQV LEKLNSPISH1740
1741 LILGYRKLSG LLAKSIQPLM ECCQADRIHG QSITYTATGR ISMTEPNLQN VAKEFSIQVG1800
1801 SDVVHISCRS PFMPTDESRC LLSADFCQLE MRILAHMSQD KALLEVMKSS QDLFIAIAAH1860
1861 WNKIEESEVT QDLRNSTKQV CYGIVYGMGM RSLAESLNCS EQEARMISDQ FHQAYKGIRD1920
1921 YTTRVVNFAR SKGFVETITG RRRYLENINS DVEHLKNQAE RQAVNSTIQG SAADIAKNAI1980
1981 LKMEKNIERY REKLALGDNS VDLVMHLHDE LIFEVPTGKA KKIAKVLSLT MENCVKLSVP2040
2041 LKVKLRIGRS WGEFKEVSV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
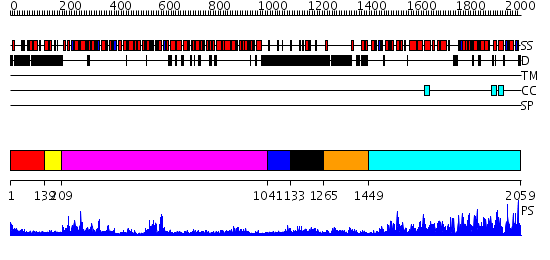
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..138] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [139..208] | 71.154902 | No description for 2db3A was found. | |
| 3 | View Details | [209..1040] | 91.39794 | No description for 2p6rA was found. | |
| 4 | View Details | [1041..1132] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [1133..1264] | 2.222997 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [1265..1448] | 3.308994 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [1449..2059] | 1000.0 | STRUCTURE OF LARGE FRAGMENT OF ESCHERICHIA COLI DNA POLYMERASE I COMPLEXED WITH D/TMP |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||
| 7 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)