













| General Information: |
|
| Name(s) found: |
Sbf-PB /
FBpp0082004
[FlyBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1973 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
intracellular signaling pathway
[IEA]
dephosphorylation [IEA] mitotic cell cycle [IMP] |
| Molecular Function: |
diacylglycerol binding
[ISS]
protein tyrosine/serine/threonine phosphatase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSRLADYFVI VGYDSDKEKT ASNVGGQPTC GKIVQRFPEK DWPDTPFIEG IEWFCQPLGW 60
61 SLSYEKQEPK FFVSVLTDID ANKHYCACLS FHETVAITQT RSVDDEDETI GSSRLLGATP 120
121 SSMDGITTTS TPASITHHSV MYAPKCLVLI SRLDCAETFK NCLGTIYTVY IENLAYGLET 180
181 LIGNILGCIQ VPPAGGPQVR FSIGAGDKQS LQPPQSSSLP TTGSGVHFLF KQLGIKNVLI 240
241 LLCSVMTENK ILFLSKCYWH LTDSCRALVA LMYPFRYTHV YIPILPAPLT EVLSTPTPFI 300
301 MGIHSSLQTE ITDLLDVIVV DLDGGLVTIP ESLTPPVPIL PSPLWEQTQD LLSMILFPNL 360
361 AQADLAFPTL ERPSAIAKTD AQIDKELRAI FMRLFAQLLQ GYRSCLTIIR IHPKPVITFH 420
421 KAGFLGARDL IESEFLFRVL DSMFFTTFVN ERGPPWRSSD AWDELYSSMN ELLKSEAQNR 480
481 NLILTHIQEL GRVLYENEGT LAHISYAQKV LRPPEGAFQR IHQPAFPRIS SEKVELIIQE 540
541 GIRKNGVPQR FHVTRNQHRI IPMGPRLPEA LDVRPNVQNS ARRLEVLRIC VSYIFENRIT 600
601 DARKLLPAVM RTLMHRDARL ILCREFFGYV HGNKAVLDHQ QFELVVRFMN KALQKSSGID 660
661 EYTVAAALLP MSTIFCRKLS TGVVQFAYTE IQDHAIWKNL QFWESTFFQD VQGQIKALYL 720
721 LHRRQNEHQK EANCVLDEVP LEEPTALEIT AEQLRKSPNI EEEKKAELAK SEESTLYSQA 780
781 IHFANRMVSL LIPLDVNVDA ASKPKPAFRL EENQSVSNSI MGSHSLSEHS DEGFEENNAL 840
841 EIGVTVGKTI SRFIDCVCTE GGVTSEHIRN LHDMVPGVVH MHIESLEPVY LEAKRHPHVQ 900
901 KPKIQTPCLL PGEDLVTDHL RCFLMPDGRE DETQCLIPAE GALFLTNYRV IFKGSPCDPL 960
961 FCEQVIVRTF PIASLLKEKK ISVLYLAHLD QTLTEGLQLR SSSFQLIKVA FDPEVTPEQI1020
1021 ESFRKILSKA RHPFDEFEYF AFQSYGTMLQ GVAPLKTKEK YSTLKGFAKK TLLRGAKKAG1080
1081 FKQKQQTKRK LVSDYDYGSA DAQETQSIDD ELEDGDEFET QNNAMPRLLT TKDVERMRER1140
1141 SYVQDWKRLG FDAESQRGFR ISNANTSYAT CRSYPAIIVA PVQCSDAAIM HLGRCFKGQR1200
1201 IPLPTWRHAN GALLIRGGQP NSKSVIGMLK NTTGSTTNAH HDVTHYPEQD KYFLALINTM1260
1261 PKLTPLALNQ YSGMNLSMSS LMGHSSSDDR QPLTPELSRK HKNNLDISDG NKSSQGGKGG1320
1321 TMKGNPKNSL AHPFRKMRLY ALGEKSQAKS NMNVDFCADF IPVDYPDIRQ SRPAFKKLIR1380
1381 ACMPSHNTNE ADGQSFAKMV EQSDWLQQIS SLMQLSGAVV DLIDLQESSV MLSLEDGSDV1440
1441 TAQLSSIAQL CLDPYYRSLD GFRVLVEKEW LAFGHRFAHR SNLKPSHANT NIAFAPTFLQ1500
1501 FLDVVHQLQR QFPMAFEFND FYLRFLAYHS VSCRFRTFLF DCELERSDSG IAAMEDKRGS1560
1561 LNAKHMFGAG GMATNGSDDE CSVYPLDIRS QRAPAPLNRI GHSIFDYIER QHNKTPIFYN1620
1621 FLYSGDKSVT LRPQNNVAAL DLWCYYTNEE LAQGAPYDLE VTTVDDEIDL SETKGKRMVI1680
1681 TAGYDNMEKC NPSAYVCLLS EVKQAETERG HLPQKWLQVW NSLEVPQLEP VARNTSLGNI1740
1741 FVQTHQHKRS TLEIIMKGRL AGYQDKYFHP HRFEKHPYTT PTNCNHCTKL LWGPVGYRCM1800
1801 DCGNSYHEKC TEHSMKNCTK YKAIDGAVGP PNVNMSQGDT ASIASSAATT ARTSSHHFYN1860
1861 QFSSNVAENR THEGHLYKRG ALLKGWKQRW FVLDSIKHQL RYYDTSEDTA PKGIIELAEV1920
1921 QSVTAAQPAQ IGAKGVDEKG FFDLKTSKRI YNFYAINANL AQEWIEKLQA CLQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
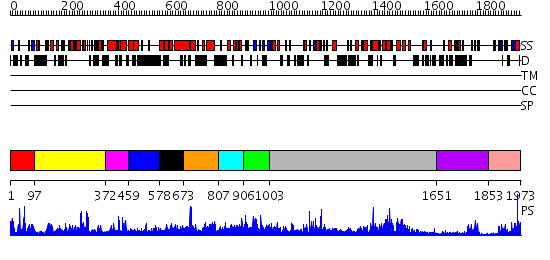
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..96] | 26.619789 | No description for PF03456.9 was found. No confident structure predictions are available. | |
| 2 | View Details | [97..371] | 62.136677 | No description for PF02141.12 was found. No confident structure predictions are available. | |
| 3 | View Details | [372..458] | 28.920819 | No description for PF03455.10 was found. No confident structure predictions are available. | |
| 4 | View Details | [459..577] | 1.158995 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [578..672] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [673..806] | 2.060955 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [807..905] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [906..1002] | 19.086186 | No description for PF02893.11 was found. No confident structure predictions are available. | |
| 9 | View Details | [1003..1650] | 142.0 | Crystal Structure of Myotubularin-related protein 2 complexed with phosphate | |
| 10 | View Details | [1651..1852] | 13.221849 | Crystal Structure of the Human Beta2-Chimaerin | |
| 11 | View Details | [1853..1973] | 20.0 | Solution Structure of the C-terminal Pleckstrin Homology Domain of Sbf1 from Mouse |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)