













| General Information: |
|
| Name(s) found: |
stau-PA /
FBpp0085962
[FlyBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1026 amino acids |
Gene Ontology: |
|
| Cellular Component: |
apical part of cell
[IDA]
cytoplasm [IDA] cell cortex [IDA] cytoplasmic mRNA processing body [IDA] apical cortex [IDA] basal cortex [IDA][NAS][TAS] microtubule associated complex [ISS] |
| Biological Process: |
intracellular mRNA localization
[NAS][TAS]
anterior/posterior axis specification, embryo [IGI] protein localization [TAS] neuroblast fate determination [TAS] pole plasm assembly [IMP][NAS] pole plasm RNA localization [IMP] RNA localization [IMP] microtubule-based process [ISS] pole plasm mRNA localization [TAS] long-term memory [IMP][TAS] regulation of pole plasm oskar mRNA localization [IGI][TAS] pole plasm protein localization [IMP][TAS] oogenesis [TAS] asymmetric protein localization involved in cell fate determination [TAS] bicoid mRNA localization [NAS][TAS] regulation of oskar mRNA translation [TAS] positive regulation of cytoplasmic mRNA processing body assembly [IMP] pole plasm oskar mRNA localization [TAS] positive regulation of oskar mRNA translation [NAS] asymmetric neuroblast division [NAS] |
| Molecular Function: |
double-stranded RNA binding
[IEA][ISS][TAS]
mRNA binding [NAS] mRNA 3'-UTR binding [IDA] microtubule binding [ISS][NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQHNVHAARP APHIRAAHHH SHSHAHMHLH PGMEQHLGPS LQQQQQPPPP PQQPPHRDLH 60
61 ARLNHHHLHA QQQQQQQTSS NQAGAVAAAG AAYHHGNINS NSGSNISSNS NQMQKIRQQH 120
121 QHLSSSNGLL GNQPPGPPPQ AFNPLAGNPA ALAYNQLPPH PPHHMAAHLG SYAAPPPHYY 180
181 MSQAKPAKYN HYGSNANSNS GSNNSNSNYA PKAILQNTYR NQKVVVPPVV QEVTPVPEPP 240
241 VTTNNATTNS TSNSTVIASE PVTQEDTSQK PETRQEPASA DDHVSTGNID ATGALSNEDT 300
301 SSSGRGGKDK TPMCLVNELA RYNKITHQYR LTEERGPAHC KTFTVTLMLG DEEYSADGFK 360
361 IKKAQHLAAS KAIEETMYKH PPPKIRRSEE GGPMRTHITP TVELNALAMK LGQRTFYLLD 420
421 PTQIPPTDSI VPPEFAGGHL LTAPGPGMPQ PPPPPAYALR QRLGNGFVPI PSQPMHPHFF 480
481 HGPGQRPFPP KFPSRFALPP PLGAHVHHGP NGPFPSVPTP PSKITLFVGK QKFVGIGRTL 540
541 QQAKHDAAAR ALQVLKTQAI SASEEALEDS MDEGDKKSPI SQVHEIGIKR NMTVHFKVLR 600
601 EEGPAHMKNF ITACIVGSIV TEGEGNGKKV SKKRAAEKML VELQKLPPLT PTKQTPLKRI 660
661 KVKTPGKSGA AAREGSVVSG TDGPTQTGKP ERRKRLNPPK DKLIDMDDAD NPITKLIQLQ 720
721 QTRKEKEPIF ELIAKNGNET ARRREFVMEV SASGSTARGT GNSKKLAKRN AAQALFELLE 780
781 AVQVTPTNET QSSEECSTSA TMSAVTAPAV EATAEGKVPM VATPVGPMPG ILILRQNKKP 840
841 AKKRDQIVIV KSNVESKEEE ANKEVAVAAE ENSNNSANSG DSSNSSSGDS QATEAASESA 900
901 LNTSTGSNTS GVSSNSSNVG ANTDGNNHAE SKNNTESSSN STSNTQSAGV HMKEQLLYLS 960
961 KLLDFEVNFS DYPKGNHNEF LTIVTLSTHP PQICHGVGKS SEESQNDAAS NALKILSKLG1020
1021 LNNAMK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
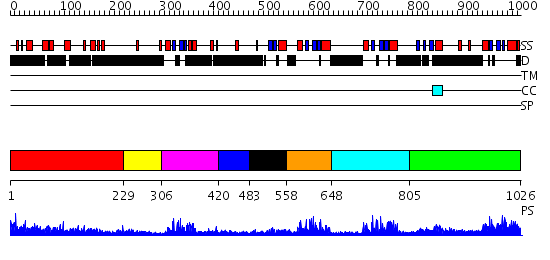
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..228] | 7.09691 | Sec24 | |
| 2 | View Details | [229..305] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [306..419] | 11.09691 | Solution structure of the first DSRM domain in Interferon-induced, double-stranded RNA-activated protein kinase | |
| 4 | View Details | [420..482] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [483..557] | 27.522879 | RNase III endonuclease catalytic domain; RNase III, C-terminal domain | |
| 6 | View Details | [558..647] | 17.522879 | Staufen, domain III | |
| 7 | View Details | [648..804] | 6.0 | Solution structure of the dsRBD from hypothetical protein BAB26260 | |
| 8 | View Details | [805..1026] | 3.24 | dsRNA-dependent protein kinase pkr |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 7 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 8 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)