













| General Information: |
|
| Name(s) found: |
Iap2-PB /
FBpp0086432
[FlyBase]
Iap2-PA / FBpp0086431 [FlyBase] |
| Description(s) found:
Found 35 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 498 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IEA]
|
| Biological Process: |
anti-apoptosis
[IDA][ISS][NAS][TAS]
defense response to Gram-negative bacterium [IMP] antimicrobial humoral response [IMP] sensory organ development [IMP] |
| Molecular Function: |
zinc ion binding
[IEA]
protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTELGMELES VRLATFGEWP LNAPVSAEDL VANGFFATGN WLEAECHFCH VRIDRWEYGD 60
61 QVAERHRRSS PICSMVLAPN HCGNVPRSQE SDNEGNSVVD SPESCSCPDL LLEANRLVTF 120
121 KDWPNPNITP QALAKAGFYY LNRLDHVKCV WCNGVIAKWE KNDNAFEEHK RFFPQCPRVQ 180
181 MGPLIEFATG KNLDELGIQP TTLPLRPKYA CVDARLRTFT DWPISNIQPA SALAQAGLYY 240
241 QKIGDQVRCF HCNIGLRSWQ KEDEPWFEHA KWSPKCQFVL LAKGPAYVSE VLATTAANAS 300
301 SQPATAPAPT LQADVLMDEA PAKEALALGI DGGVVRNAIQ RKLLSSGCAF STLDELLHDI 360
361 FDDAGAGAAL EVREPPEPSA PFIEPCQATT SKAASVPIPV ADSIPAKPQA AEAVANISKI 420
421 TDEIQKMSVA TPNGNLSLEE ENRQLKDARL CKVCLDEEVG VVFLPCGHLA TCNQCAPSVA 480
481 NCPMCRADIK GFVRTFLS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
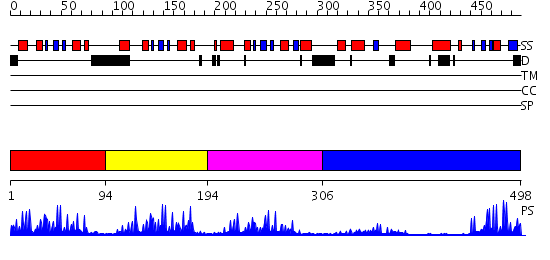
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..93] | 27.522879 | Structure of an ML-IAP/XIAP chimera bound to a 9mer peptide derived from Smac | |
| 2 | View Details | [94..193] | 24.0 | BIR2 domain of DIAP1 | |
| 3 | View Details | [194..305] | 23.39794 | 2MIHB/C-IAP-1 | |
| 4 | View Details | [306..498] | 8.39794 | Cbl; N-terminal domain of cbl (N-cbl); CBL |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)