













| General Information: |
|
| Name(s) found: |
Pka-R2-PB /
FBpp0087524
[FlyBase]
Pka-R2-PA / FBpp0087523 [FlyBase] |
| Description(s) found:
Found 47 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 377 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA]
cAMP-dependent protein kinase complex [IEA][ISS][NAS] |
| Biological Process: |
behavioral response to cocaine
[IMP][NAS]
locomotor rhythm [IMP][NAS] signal transduction [IEA] regulation of protein amino acid phosphorylation [IEA] axon guidance [IMP] behavioral response to ethanol [IMP][NAS][TAS] protein amino acid phosphorylation [ISS] circadian rhythm [IMP][NAS] rhythmic behavior [TAS] |
| Molecular Function: |
protein binding
[IPI]
cAMP-dependent protein kinase regulator activity [ISS][NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSDSSRRIQ VPEELKEVLL QFSISFLVEQ PPDVIDYAVE YFTKLQSERP SVSHTDQSTD 60
61 DQLSVNSQDA DAEPPVMASS RRKSVFAEAY DPEADDDDDG ATAVFPKTDE QRARLVESVK 120
121 NVLLFRSLEK EQMNQVLDAM FERKVQPGDF IIRQGDDGDN FYVIESGVYK VYINDKHINT 180
181 YNHTGLFGEL ALLYNMPRAA TVQAETSGLL WAMDRQTFRR ILLKSAFRKR KMYEELLNSV 240
241 PMLKALQNYE RMNLADALVS KSYDNGERII KQGDAADGMY FIEEGTVSVR MDQDDAEVEI 300
301 SQLGKGQYFG ELALVTHRPR AASVYATGGV VKLAFLDVKA FERLLGPCMD IMKRNIDDYE 360
361 SQLVKIFGSK NNITDTR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
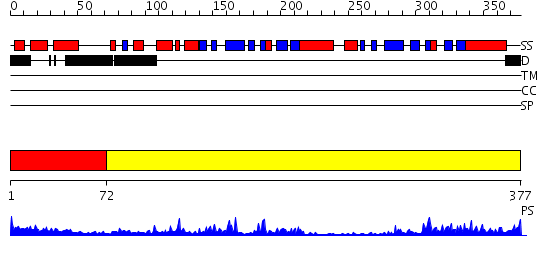
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..71] | 3.18 | No description for 2izyA was found. | |
| 2 | View Details | [72..377] | 51.0 | Regulatory subunit of Protein kinase A |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)