













| General Information: |
|
| Name(s) found: |
UBXN1_MOUSE
[Swiss-Prot]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Mus musculus |
| Length: | 297 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAELTALESL IEMGFPRGRA EKALALTGNQ GIEAAMDWLM EHEDDPDVDE PLETPLSHVL 60
61 GREPTPSEQV GPEGSGSAAG ESRPILTEEE RQEQTKRMLE LVAQKQRERE EREEREALER 120
121 EKQRRRQGQE LSVARQKLQE DEMRRAAEER RREKAEELAA RQRVREKIER DKAERAKKYG 180
181 GSVGSRSSPP ATDPGPVPSS PSQEPPTKRE YDQCRIQVRL PDGTSLTQTF RAREQLAAVR 240
241 LYVELHRGEE PGQDQDPVQL LSGFPRRAFS EADMERPLQE LGLVPSAVLI VAKKCPS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
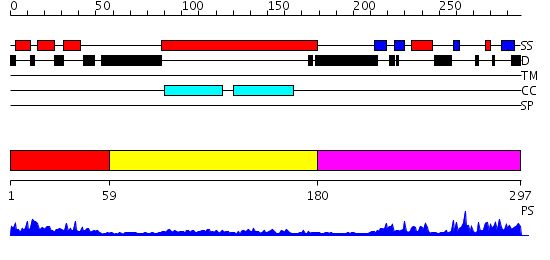
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..58] | 14.30103 | Solution Structure of RSGI RUH-027, a UBA domain from Mouse cDNA | |
| 2 | View Details | [59..179] | 1.01 | No description for 2j63A was found. | |
| 3 | View Details | [180..297] | 18.39794 | Crystal structure of AAA ATPase p97/VCP ND1 in complex with p47 C |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)