













| General Information: |
|
| Name(s) found: |
SNF8 /
YPL002C
[SGD]
|
| Description(s) found:
Found 38 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 233 amino acids |
Gene Ontology: |
|
| Cellular Component: |
ESCRT II complex
[IDA |
| Biological Process: |
ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway
[IC telomere maintenance [IMP negative regulation of transcription from RNA polymerase II promoter by glucose [IMP protein targeting to vacuole [IMP |
| Molecular Function: |
protein binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKQFGLAAFD ELKDGKYNDV NKTILEKQSV ELRDQLMVFQ ERLVEFAKKH NSELQASPEF 60
61 RSKFMHMCSS IGIDPLSLFD RDKHLFTVND FYYEVCLKVI EICRQTKDMN GGVISFQELE 120
121 KVHFRKLNVG LDDLEKSIDM LKSLECFEIF QIRGKKFLRS VPNELTSDQT KILEICSILG 180
181 YSSISLLKAN LGWEAVRSKS ALDEMVANGL LWIDYQGGAE ALYWDPSWIT RQL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
SNF8 VPS36 |
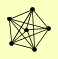
| View Details | Krogan NJ, et al. (2006) |
|
ADH7 HCM1 OLA1 SNF8 VPS25 VPS36 |
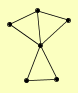
| View Details | Qiu et al. (2008) |
|
SNF7 SNF8 SRN2 STP22 VPS25 VPS36 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
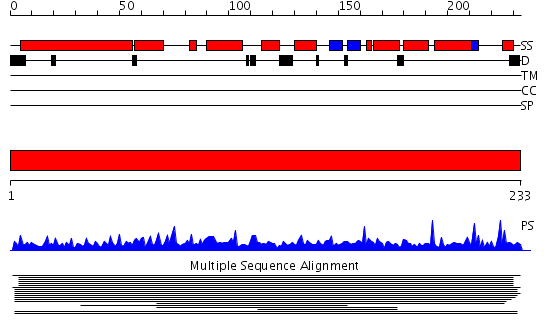
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..233] | 3.012982 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)