













| General Information: |
|
| Name(s) found: |
IOC4 /
YMR044W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 475 amino acids |
Gene Ontology: |
|
| Cellular Component: |
ISW1 complex
[IPI |
| Biological Process: |
chromatin remodeling
[IPI |
| Molecular Function: |
protein binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSEAIFQPTD IVLAKVKGFS AWPAMIIPNE LIPDNILKTK PVSVHKGKSG SDKKANEDID 60
61 ADMESEARDR EQSEEEEDIE DFGESEANPE KFIIYTPVLK FRKNDTLKST YCVKFFCDDS 120
121 YIWVKPMDMK ILTSEDCRKW LSGKQRKNKK LIPAYEMAMR GKNGIDIWEF VEYGSYGKPD 180
181 EEEYVEEEEE ENEPEKKAIR PTRSSSRQRQ KRASETEKSE GGNSNKRKRV TRSTRQQAID 240
241 ASEEEEEEEE EKVQEAVRKR PQRTKTKKVV VSKTKPNPKT KAKKEKPKPP KPIKYHFEDD 300
301 EDWSIVGLGP QDLSIEKTMD PIAKKLSQKK NLEKHVEIKL DLEDKLAGIN KLLCDVLCSA 360
361 INQAVSIKDD FEIILDELQI ALDTRGSRNE FITIFQSNNS LLLNFRILFN LRKRELNKWD 420
421 LWDRFQDIFK HIYSYQFIPD TEDWQLEQNM EIEEMDREKP SFSEDVKEEE SKVGA |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
IOC2 IOC4 |
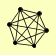
| View Details | Krogan NJ, et al. (2006) |
|
ESC8 IOC2 IOC3 IOC4 ISW1 YJL068C |
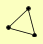
| View Details | Qiu et al. (2008) |
|
IOC2 IOC3 IOC4 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
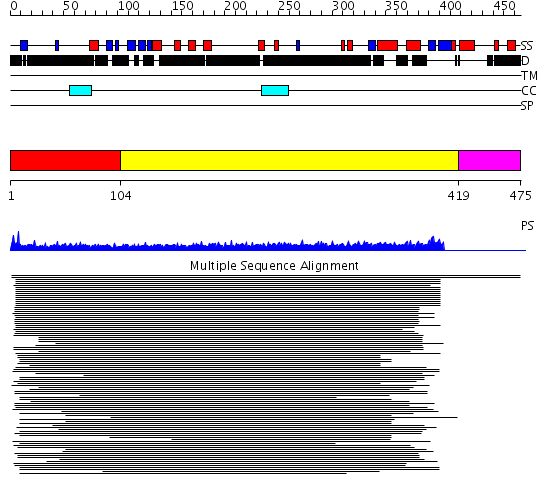
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..103] | 23.638272 | PWWP domain No confident structure predictions are available. | |
| 2 | View Details | [104..418] | 17.242997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [419..475] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)