













| General Information: |
|
| Name(s) found: |
CDC5 /
YMR001C
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 705 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cellular bud neck [IDA spindle pole [IDA |
| Biological Process: |
DNA-dependent DNA replication
[IDA protein amino acid phosphorylation [IDA |
| Molecular Function: |
protein kinase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLGPLKAIN DKQLNTRSKL VHTPIKGNTA DLVGKENHFK QTKRLDPNND HHHQPAQKKK 60
61 REKLSALCKT PPSLIKTRGK DYHRGHFLGE GGFARCFQIK DDSGEIFAAK TVAKASIKSE 120
121 KTRKKLLSEI QIHKSMSHPN IVQFIDCFED DSNVYILLEI CPNGSLMELL KRRKVLTEPE 180
181 VRFFTTQICG AIKYMHSRRV IHRDLKLGNI FFDSNYNLKI GDFGLAAVLA NESERKYTIC 240
241 GTPNYIAPEV LMGKHSGHSF EVDIWSLGVM LYALLIGKPP FQARDVNTIY ERIKCRDFSF 300
301 PRDKPISDEG KILIRDILSL DPIERPSLTE IMDYVWFRGT FPPSIPSTVM SEAPNFEDIP 360
361 EEQSLVNFKD CMEKSLLLES MSSDKIQRQK RDYISSIKSS IDKLEEYHQN RPFLPHSLSP 420
421 GGTKQKYKEV VDIEAQRRLN DLAREARIRR AQQAVLRKEL IATSTNVIKS EISLRILASE 480
481 CHLTLNGIVE AEAQYKMGGL PKSRLPKIKH PMIVTKWVDY SNKHGFSYQL STEDIGVLFN 540
541 NGTTVLRLAD AEEFWYISYD DREGWVASHY LLSEKPRELS RHLEVVDFFA KYMKANLSRV 600
601 STFGREEYHK DDVFLRRYTR YKPFVMFELS DGTFQFNFKD HHKMAISDGG KLVTYISPSH 660
661 ESTTYPLVEV LKYGEIPGYP ESNFREKLTL IKEGLKQKST IVTVD |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | #34 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #14 Mitotic Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #16 Mitotic-New search criteria | Keck JM, et al. (2011) | |
| View Run | #15 Mitotic 2nd-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #18 Mitotic Prep4-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
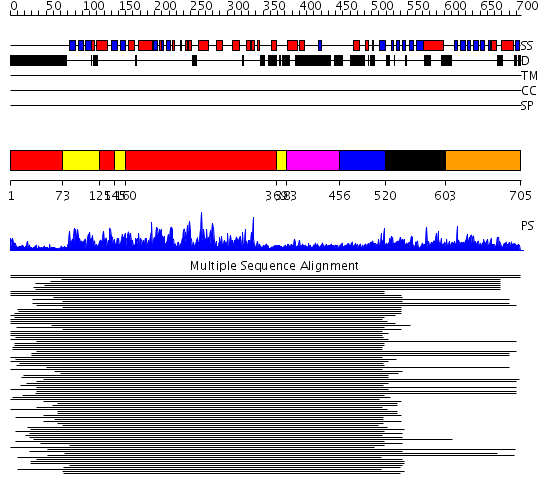
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..72] [125..144] [160..368] |
486.0 | cAMP-dependent PK, catalytic subunit | |
| 2 | View Details | [73..124] [145..159] [369..382] |
486.0 | cAMP-dependent PK, catalytic subunit | |
| 3 | View Details | [383..455] | 17.69897 | Calmodulin | |
| 4 | View Details | [456..519] | 17.69897 | Calmodulin | |
| 5 | View Details | [520..602] | 8.38 | Sak kinase Polo domain | |
| 6 | View Details | [603..705] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)