













| General Information: |
|
| Name(s) found: |
ITT1 /
YML068W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 464 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
regulation of translational termination
[IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALTQFENDL EILRDMYPEL EMKSVKVEEE GEFPQRINGK LLFKISLLAD VNIEFGEQHM 60
61 LLSNLSNECV EFTIYSCHYP DIRRCVVMDI KSLWISTDEK KMLIDKALRL VEETVDMSIE 120
121 FADSFTSILI LIFGFLIDDT AILLFPNGIR KCLTQDQYDL FKQISEEATL QKVSRSNYHC 180
181 CICMEMEKGV RMIKLPCENA NVEHYLCRGC AKSYFTAMIQ ENRISSVRCP QCEYKELKLE 240
241 DFKSYKKMLK ALFTPLIPVS FLKEVIDTEL CERYEKMFYN QAATRLSKYC PYACVTCRRC 300
301 DSWCTKEDLD DAMIQCQKCH FVFCFDCLHA WHGYNNKCGK KVSLSTDIIE EYLDDTVTSY 360
361 ERKRKLEAKY GRRVLELEVN DYLAEKMLDL AIKKEGSNLQ RCPKCKVVVE RSEGCNKMKC 420
421 EVCGTLFCFI CGVLLYPEDP YEHFREAYSG CYGRLFEGMP GTET |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
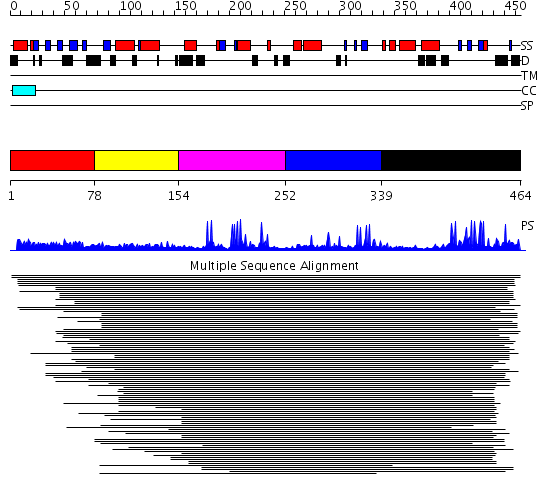
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..77] | 2.045993 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [78..153] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [154..251] | 7.120997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [252..338] | 18.920819 | IBR domain No confident structure predictions are available. | |
| 5 | View Details | [339..464] | 2.075721 | IBR domain No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.81 |
Source: Reynolds et al. (2008)