













| General Information: |
|
| Name(s) found: |
PIF1 /
YML061C
[SGD]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 859 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC mitochondrion [IDA |
| Biological Process: |
DNA recombination
[IMP telomere maintenance [IMP mitochondrial genome maintenance [IMP |
| Molecular Function: |
DNA helicase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPKWIRSTLN HIIPRRPFIC SFNSFLLLKN VSHAKLSFSM SSRGFRSNNF IQAQLKHPSI 60
61 LSKEDLDLLS DSDDWEEPDC IQLETEKQEK KIITDIHKED PVDKKPMRDK NVMNFINKDS 120
121 PLSWNDMFKP SIIQPPQLIS ENSFDQSSQK KSRSTGFKNP LRPALKKESS FDELQNNSIS 180
181 QERSLEMINE NEKKKMQFGE KIAVLTQRPS FTELQNDQDD SNLNPHNGVK VKIPICLSKE 240
241 QESIIKLAEN GHNIFYTGSA GTGKSILLRE MIKVLKGIYG RENVAVTAST GLAACNIGGI 300
301 TIHSFAGIGL GKGDADKLYK KVRRSRKHLR RWENIGALVV DEISMLDAEL LDKLDFIARK 360
361 IRKNHQPFGG IQLIFCGDFF QLPPVSKDPN RPTKFAFESK AWKEGVKMTI MLQKVFRQRG 420
421 DVKFIDMLNR MRLGNIDDET EREFKKLSRP LPDDEIIPAE LYSTRMEVER ANNSRLSKLP 480
481 GQVHIFNAID GGALEDEELK ERLLQNFLAP KELHLKVGAQ VMMVKNLDAT LVNGSLGKVI 540
541 EFMDPETYFC YEALTNDPSM PPEKLETWAE NPSKLKAAME REQSDGEESA VASRKSSVKE 600
601 GFAKSDIGEP VSPLDSSVFD FMKRVKTDDE VVLENIKRKE QLMQTIHQNS AGKRRLPLVR 660
661 FKASDMSTRM VLVEPEDWAI EDENEKPLVS RVQLPLMLAW SLSIHKSQGQ TLPKVKVDLR 720
721 RVFEKGQAYV ALSRAVSREG LQVLNFDRTR IKAHQKVIDF YLTLSSAESA YKQLEADEQV 780
781 KKRKLDYAPG PKYKAKSKSK SNSPAPISAT TQSNNGIAAM LQRHSRKRFQ LKKESNSNQV 840
841 HSLVSDEPRG QDTEDHILE |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
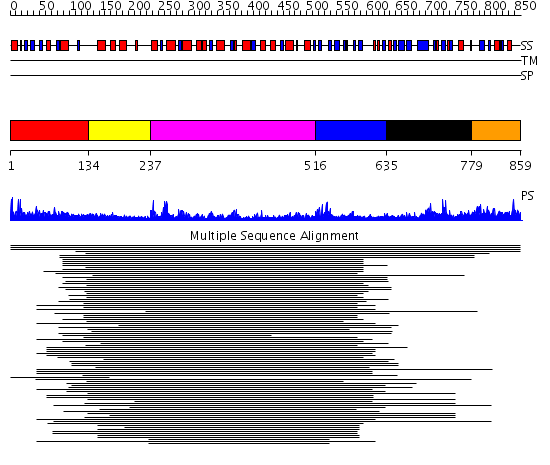
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..133] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [134..236] | 3.221849 | Signal recognition particle receptor, FtsY; GTPase domain of the signal recognition particle receptor FtsY | |
| 3 | View Details | [237..515] | 37.69897 | DEXX box DNA helicase | |
| 4 | View Details | [516..634] | 37.69897 | DEXX box DNA helicase | |
| 5 | View Details | [635..778] | 26.132995 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [779..859] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|