













| General Information: |
|
| Name(s) found: |
SPO14 /
YKR031C
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1683 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endosome
[IDA prospore membrane [IDA |
| Biological Process: |
ascospore-type prospore formation
[IMP phospholipid metabolic process [IDA exocytosis [IGI |
| Molecular Function: |
phosphatidylinositol-3-phosphate binding
[IDA phospholipase D activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNVSTASGT HFAPPQADRS VTEEVDRVNS RPDELENQEV LRQLPENGNL TSSLQREKRR 60
61 TPNGKEAERK HALPKSFVDR NLSDVSPNHS LDHIMHSNEH DPRRGSDEEN MHRLYNNLHS 120
121 SNNNVHSKRN SKREEERAPQ RRSSSVAYTQ QQFNGWKKEF GHAFKKISAI GRLKSSVNSP 180
181 TPAGSGHRHN QHQHQQVNEE DLYTQRLASD LLDSLLAGCP ASLFASTQFL RDEHGKRRAP 240
241 LLLAKLDVRV SPLKNDNNIL DITNSNHNHR GNNNNNTGEN SDRRPSIPRS SSIISISSNV 300
301 AEFMYSRNEN SLFRIHLEYG IDEDRLKWSI IRSYKDIKSL HHKLKIVAFQ QLTISKLYSD 360
361 NNRYHSLQLP HFPHYKEMVK ERNVMEKKAE NKPSSAASAP HTSENNNNDN GSNITSLETL 420
421 SSSEISEFNI DNVKMKHLQD LIDEPDDFSQ PIHLRLERYL RLLNIALCLR PHANRLFEFY 480
481 ELSPLGNLLS RESGFQGKQG YLVIRSTAKA QGWRVSHFGK HAFKDMIDRH TTKWFLVRNS 540
541 YLTYVSDLSS TTPLDVFLID WKFKVRFSGN KNNILDNENE INWIIHDPNL EINDELEEFG 600
601 IENDANNILD KNGKSKTHQK KSNISSKLLL LTLENSERKL KIICKSESSL KQWMSSIIKM 660
661 STSTPWSKPN RFGSFAPVRT NSFCKFLVDG RDYFWSLSEA LLMAKDVIYI HDWWLSPELY 720
721 LRRPVKGNQG FRIDRMLKSC AEKGIKIFIV IYRNVGNIVG TDSLWTKHSM LNLHPNIHII 780
781 RSPNQWLQNT YFWAHHEKFV VIDETFAFIG GTDLCYGRYD TFEHVLRDDA ESLLDQNFPG 840
841 KDYSNARIAD FHDLDKPFES MYDRKVIPRM PWHDVQMMTL GEPARDLARH FVQRWNYLLR 900
901 AKRPSRLTPL LTPPSDLTAE ELKSLPMFEI LREKSTCETQ ILRSAGNWSL GLKETECSIQ 960
961 NAYLKLIEQS EHFIYIENQF FITSTVWNGT CVLNKIGDAL VDRIVKANQE KKPWKAFILI1020
1021 PLMPGFDSPV DTAEASSLRL IMQFQYQSIS RGEHSTFSKL KKLNIDPAQY IQFFSLRKWS1080
1081 TFAPNERLIT EQLYVHAKIL IADDRRCIIG SANINERSQL GNRDSEVAIL IRDTDLIKTK1140
1141 MNGDDYYAGK FPWELRQRLM REHLGCDVDL VEFVEKKFER FEKFAAKNYE KLHTLSKEGD1200
1201 SGNNWSDREM IDSAMIELGY REIFGCKFSP QWKSGHGNSV DDGSTQCGIN EKEVGREDEN1260
1261 VYEKFFNSVD YGKSSRKRTP LPKHNFASLG LTFNHRAGIE NVGIRDHKVL STDPRLRKND1320
1321 EHKKEVDGYG PDCWKKESNK KFKADATEQL KEWALNSLAS KVLDDKEMIK SEIPEGFSNY1380
1381 LPNEKDLEMY LTDKTVTNRN KWSMLKRICY LQYLSHKLDE RKTQRLKKIK DMRRHLSSST1440
1441 ESTRNGSNSL PLNEKSNEGE STNVDQDIEG DEYHRLHEDI LKNQELDDGS LDDLLSQIIP1500
1501 KITNFNSGEI DDAKKEELLK LNFIDPYSFE DPLISSFSEG LWFTIALRNT LLYKLVFHCQ1560
1561 PDNAVQNWKE YGEFTELEQE FQINQEKLID LEAENINSTT TNVVDKDREK EKMRKAAELR1620
1621 MKLSGSLLYG FNQKVFDKHT AQRILERIHG HLVIFPTEWL AKEVESRNWI FNSDRLSPME1680
1681 IYN |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Gavin AC, et al. (2006) |

|
SPO14 YGL036W |
| View Details | Gavin AC, et al. (2002) |
|
MUM2 SPO14 YGL036W |
| View Details | Qiu et al. (2008) |

|
GIP1 SPO14 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Toshima J, et al (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
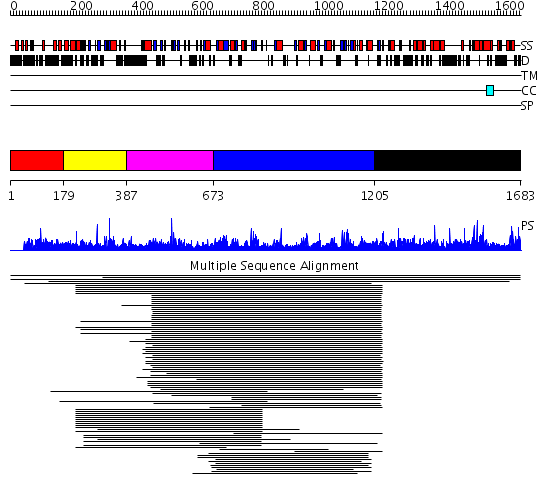
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..178] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [179..386] | 1.012992 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [387..672] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [673..1204] | 24.69897 | Phospholipase D | |
| 5 | View Details | [1205..1683] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1227..1511] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1512..1683] | 1.010978 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)