













| General Information: |
|
| Name(s) found: |
MET3 /
YJR010W
[SGD]
|
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 511 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA mitochondrion [IDA |
| Biological Process: |
sulfate assimilation
[TAS methionine metabolic process [IMP |
| Molecular Function: |
sulfate adenylyltransferase (ATP) activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPAPHGGILQ DLIARDALKK NELLSEAQSS DILVWNLTPR QLCDIELILN GGFSPLTGFL 60
61 NENDYSSVVT DSRLADGTLW TIPITLDVDE AFANQIKPDT RIALFQDDEI PIAILTVQDV 120
121 YKPNKTIEAE KVFRGDPEHP AISYLFNVAG DYYVGGSLEA IQLPQHYDYP GLRKTPAQLR 180
181 LEFQSRQWDR VVAFQTRNPM HRAHRELTVR AAREANAKVL IHPVVGLTKP GDIDHHTRVR 240
241 VYQEIIKRYP NGIAFLSLLP LAMRMSGDRE AVWHAIIRKN YGASHFIVGR DHAGPGKNSK 300
301 GVDFYGPYDA QELVESYKHE LDIEVVPFRM VTYLPDEDRY APIDQIDTTK TRTLNISGTE 360
361 LRRRLRVGGE IPEWFSYPEV VKILRESNPP RPKQGFSIVL GNSLTVSREQ LSIALLSTFL 420
421 QFGGGRYYKI FEHNNKTELL SLIQDFIGSG SGLIIPNQWE DDKDSVVGKQ NVYLLDTSSS 480
481 ADIQLESADE PISHIVQKVV LFLEDNGFFV F |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
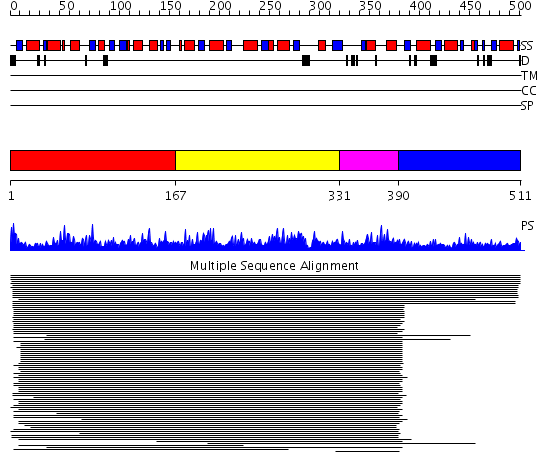
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..166] | 11000.0 | ATP sulfurylase N-terminal domain; ATP sulfurylase central domain; ATP sulfurylase C-terminal domain | |
| 2 | View Details | [167..330] | 11000.0 | ATP sulfurylase N-terminal domain; ATP sulfurylase central domain; ATP sulfurylase C-terminal domain | |
| 3 | View Details | [331..389] | 11000.0 | ATP sulfurylase N-terminal domain; ATP sulfurylase central domain; ATP sulfurylase C-terminal domain | |
| 4 | View Details | [390..511] | 11000.0 | ATP sulfurylase N-terminal domain; ATP sulfurylase central domain; ATP sulfurylase C-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)