













| General Information: |
|
| Name(s) found: |
SPT10 /
YJL127C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 640 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[TAS |
| Biological Process: |
DNA repair
[IMP chromatin remodeling [TAS regulation of transcription from RNA polymerase II promoter, global [TAS |
| Molecular Function: |
histone acetyltransferase activity
[IDA sequence-specific DNA binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLNQHTSSVP DDEHLQMAHQ NSSSEVRNEA AVPDQLLTPL QPYTILLKDG ETIATMYPIP 60
61 AYPDLLPLGL LNFLLDEFNM EVEKGDSFPY YETLSLEEFK NVWFHNDGHV CIMVLGEIPE 120
121 LDYSMDTEAD TNDNFGTEIE TTKHTTQYKK RKERRNLNLS MQWEKQCLGI FDLKPAYPGR 180
181 SAHVVTGTFL VNAGIRGKGI GKTLMETFIE WSKKLGFTSS FFPLIYGTNV GIRRILEGLN 240
241 FRRIGKLPEA GILKGFDVPV DSFMYGKEFT HITKSIDLLR DPQKSIEIGK YERLKHFLET 300
301 GKYPLHCDRN EKARLRVLSK THSVLNGKLM TKGKEIIYDT DQQIQIALEI HLMEHLGINK 360
361 VTSKIGEKYH WRGIKSTVSE VISRCQKCKM RYKDGTGVII EQKRAVKQAH MLPTQHIETI 420
421 NNPRKSKKHD NALLGQAINF PQNIISSTLN DVEGEPTPPD TNIVQPTFQN ATNSPATTAE 480
481 ANEANKRSEF LSSIQSTPLL DDEQSMNSFN RFVEEENSRK RRKYLDVASN GIVPHLTNNE 540
541 SQDHANPVNR DERDMNHSVP DLDRNDHTIM NDAMLSLEDN VMAALEMVQK EQQQKINHRG 600
601 EDVTGQQIDL NNSEGNENSV TKIVNNESNT FTEHNSNIYY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
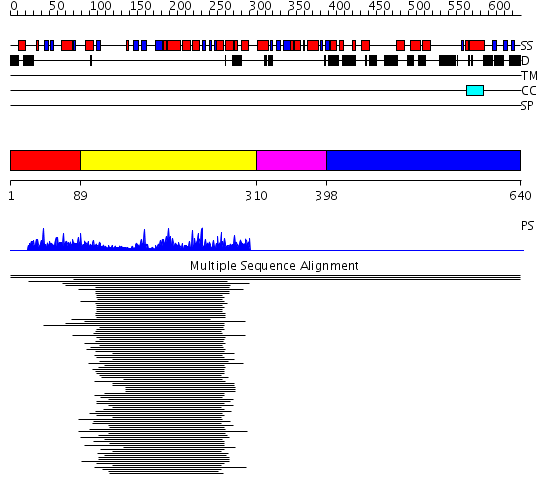
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..88] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [89..309] | 7.221849 | Aminoglycoside 6'-N-acetyltransferase | |
| 3 | View Details | [310..397] | 8.53 | N-terminal Zn binding domain of HIV integrase; Retroviral integrase, catalytic domain | |
| 4 | View Details | [398..640] | 8.53 | N-terminal Zn binding domain of HIV integrase; Retroviral integrase, catalytic domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)