













| General Information: |
|
| Name(s) found: |
ARG3 /
YJL088W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 338 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA |
| Biological Process: |
arginine biosynthetic process
[TAS ornithine metabolic process [TAS |
| Molecular Function: |
ornithine carbamoyltransferase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTTASTPSS LRHLISIKDL SDEEFRILVQ RAQHFKNVFK ANKTNDFQSN HLKLLGRTIA 60
61 LIFTKRSTRT RISTEGAATF FGAQPMFLGK EDIQLGVNES FYDTTKVVSS MVSCIFARVN 120
121 KHEDILAFCK DSSVPIINSL CDKFHPLQAI CDLLTIIENF NISLDEVNKG INSKLKMAWI 180
181 GDANNVINDM CIACLKFGIS VSISTPPGIE MDSDIVDEAK KVAERNGATF ELTHDSLKAS 240
241 TNANILVTDT FVSMGEEFAK QAKLKQFKGF QINQELVSVA DPNYKFMHCL PRHQEEVSDD 300
301 VFYGEHSIVF EEAENRLYAA MSAIDIFVNN KGNFKDLK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
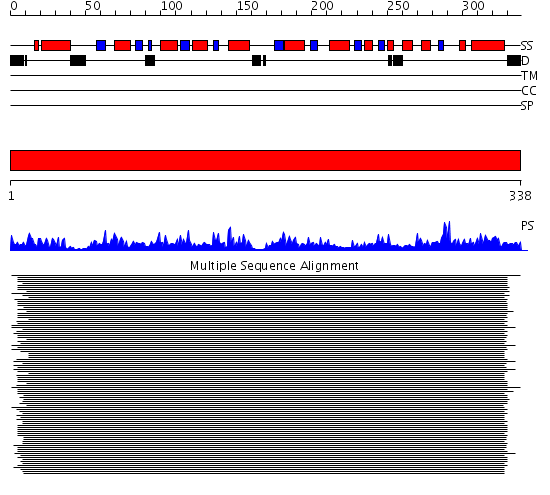
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..338] | 115.0 | Refined Structure of Pyrococcus furiosus Ornithine Carbamoyltransferase at 1.87 A |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)