













| General Information: |
|
| Name(s) found: |
HOP1 /
YIL072W
[SGD]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 605 amino acids |
Gene Ontology: |
|
| Cellular Component: |
lateral element
[IPI condensed nuclear chromosome [IDA |
| Biological Process: |
synaptonemal complex assembly
[IMP synapsis [IMP |
| Molecular Function: |
four-way junction DNA binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNKQLVKPK TETKTEITTE QSQKLLQTML TMSFGCLAFL RGLFPDDIFV DQRFVPEKVE 60
61 KNYNKQNTSQ NNSIKIKTLI RGKSAQADLL LDWLEKGVFK SIRLKCLKAL SLGIFLEDPT 120
121 DLLENYIFSF DYDEENNVNI NVNLSGNKKG SKNADPENET ISLLDSRRMV QQLMRRFIII 180
181 TQSLEPLPQK KFLTMRLMFN DNVDEDYQPE LFKDATFDKR ATLKVPTNLD NDAIDVGTLN 240
241 TKHHKVALSV LSAATSSMEK AGNTNFIRVD PFDLILQQQE ENKLEESVPT KPQNFVTSQT 300
301 TNVLGNLLNS SQASIQPTQF VSNNPVTGIC SCECGLEVPK AATVLKTCKS CRKTLHGICY 360
361 GNFLHSSIEK CFTCIFGPSL DTKWSKFQDL MMIRKVFRFL VRKKKGFPAS ITELIDSFIN 420
421 VEDQNNEVKE RVAFALFVFF LDETLCLDNG GKPSQTIRYV TSSVLVDVKG IVIPNTRKQL 480
481 NVNHEYKWHF TTSSPKAESF YQEVLPNSRK QVESWLQDIT NLRKVYSEAL SPSSTLQELD 540
541 LNSSLPTQDP IISGQKRRRY DLDEYLEEDK SSVVNDTIKA KDFDESVPAK IRKISVSKKT 600
601 LKSNW |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |
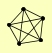
|
HOP1 RED1 TOP2 ZIP1 ZIP2 |
| View Details | Qiu et al. (2008) |
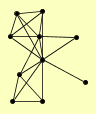
|
DMC1 HOP1 HOP2 MND1 NDJ1 RED1 SPO13 TOP2 ZIP1 ZIP2 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
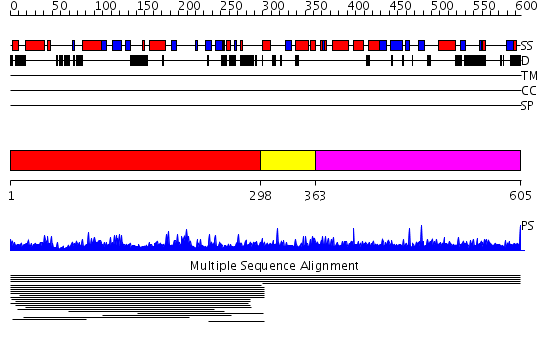
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..297] | 44.0 | The spindle assembly checkpoint protein mad2 | |
| 2 | View Details | [298..362] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [363..605] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.76 |
Source: Reynolds et al. (2008)