













| General Information: |
|
| Name(s) found: |
ZIP1 /
YDR285W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 875 amino acids |
Gene Ontology: |
|
| Cellular Component: |
transverse filament
[IDA |
| Biological Process: |
synapsis
[IGI |
| Molecular Function: |
SUMO polymer binding
[IPI chromatin binding [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNFFRDSSM GFKPRPNIFA KLRVRDVDSD SSANTVVENS SNCLDVGSSI EGDDTFKKPH 60
61 KTSTEQELIT SMSLSQRNHG YSDDMEIGSP KKTTSTDQYN RILKNDVAAI ENDTDEDFEI 120
121 TEVREVSEGV AKETKESHGD PNDSETTLKD SKMHEYTMTN GKAPLHTSIN NSSTSSNDVL 180
181 LEAFTNTQRI CSNLKQELQK QQQDNAKLKV RLQSYASNSD KINEKVGKYK SCLETLQERI 240
241 ATLTSHKNNQ ETKLKDLRQN HQLYQRRISG FKTSIENLNK TINDLGKNKK EADAELMKKG 300
301 KEIEYLKREL DDCSGQLSEE KIKNSSLIQE MGKNREEMIK SIENFFSEDK AHHLLQFNKF 360
361 EERVHDLFEK KLQKHFDVAK DTLNVGLRNT TVELSSNTET MLKQQYEDIK ENLEQKMSSS 420
421 KDEMAKTINE LSVTQKGLIM GVQEELLTSS GNIQTALVSE MNNTRQELLD DASQTAKNYA 480
481 SLENLVKAYK AEIVQSNEYE ERIKHLESER STLSSQKNQI ISSLGTKEAQ YEDLVKKLEA 540
541 KNIEISQISG KEQSLTEKNE NLSNELKKVQ DQLEKLNNLN ITTKSNYENK ISSQNEIVKA 600
601 LVSENDTLKQ RIQQLVEIKE NEQKDHTTKL EAFQKNNEQL QKLNVEVVQL KAHELELEEQ 660
661 NRHLKNCLEK KETGVEESLS DVKTLKQQVI VLKSEKQDIT AEKLELQDNL ESLEEVTKNL 720
721 QQKVQSQKRE LEQKIKELEE IKNHKRNEPS KKGTQNFTKP SDSPKKNATT SNLFPNNSAA 780
781 IHSPMKKCPK VDHISKSRIN SSKETSKFND EFDLSSSSND DLELTNPSPI QIKPVRGKIK 840
841 KGSNCMKPPI SSRKKLLLVE DEDQSLKISK KRRRK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
HOP1 RED1 TOP2 ZIP1 ZIP2 |
| View Details | Qiu et al. (2008) |
|
HOP1 HSK3 RED1 ZIP1 ZIP2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
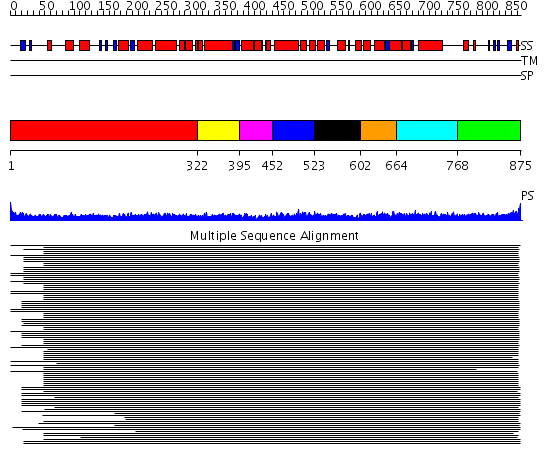
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..321] | 54.9897 | Heavy meromyosin subfragment | |
| 2 | View Details | [322..394] | 9.69897 | Tropomyosin | |
| 3 | View Details | [395..451] | N/A | Confident ab initio structure predictions are available. | |
| 4 | View Details | [452..522] | 12.0 | Tropomyosin | |
| 5 | View Details | [523..601] | 12.0 | Tropomyosin | |
| 6 | View Details | [602..663] | 12.0 | Tropomyosin | |
| 7 | View Details | [664..767] | 12.0 | Tropomyosin | |
| 8 | View Details | [768..875] | 1.763994 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)