













| General Information: |
|
| Name(s) found: |
IME4 /
YGL192W
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 600 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
meiosis
[IMP mRNA modification [IMP |
| Molecular Function: |
mRNA methyltransferase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MINDKLVHFL IQNYDDILRA PLSGQLKDVY SLYISGGYDD EMQKLRNDKD EVLQFEQFWN 60
61 DLQDIIFATP QSIQFDQNLL VADRPEKIVY LDVFSLKILY NKFHAFYYTL KSSSSSCEEK 120
121 VSSLTTKPEA DSEKDQLLGR LLGVLNWDVN VSNQGLPREQ LSNRLQNLLR EKPSSFQLAK 180
181 ERAKYTTEVI EYIPICSDYS HASLLSTSVY IVNNKIVSLQ WSKISACQEN HPGLIECIQS 240
241 KIHFIPNIKP QTDISLGDCS YLDTCHKLNT CRYIHYLQYI PSCLQERADR ETASENKRIR 300
301 SNVSIPFYTL GNCSAHCIKK ALPAQWIRCD VRKFDFRVLG KFSVVIADPA WNIHMNLPYG 360
361 TCNDIELLGL PLHELQDEGI IFLWVTGRAI ELGKESLNNW GYNVINEVSW IKTNQLGRTI 420
421 VTGRTGHWLN HSKEHLLVGL KGNPKWINKH IDVDLIVSMT RETSRKPDEL YGIAERLAGT 480
481 HARKLEIFGR DHNTRPGWFT IGNQLTGNCI YEMDVERKYQ EFMKSKTGTS HTGTKKIDKK 540
541 QPSKLQQQHQ QQYWNNMDMG SGKYYAEAKQ NPMNQKHTPF ESKQQQKQQF QTLNNLYFAQ 600
601 |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Ho Y, et al. (2002) |
|
CAT5 IME4 PRP28 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
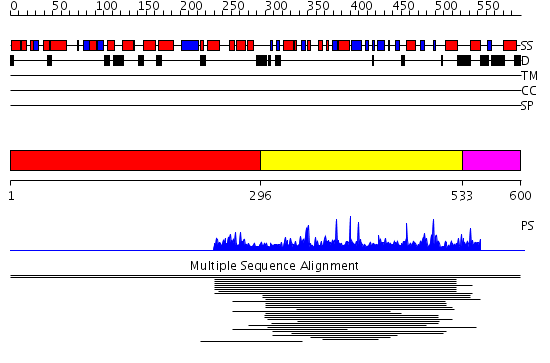
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..295] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [296..532] | 6.024967 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [533..600] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)