













| General Information: |
|
| Name(s) found: |
PKC1 /
YBL105C
[SGD]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1151 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoskeleton [IDA site of polarized growth [IDA cytoplasm [IDA |
| Biological Process: |
signal transduction
[IMP cellular cell wall organization [IMP actin filament organization [IGI intracellular protein kinase cascade [IMP protein amino acid phosphorylation [IDA |
| Molecular Function: |
protein kinase C activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSFSQLEQNI KKKIAVEENI IRGASALKKK TSNVMVIQKC NTNIREARQN LEYLEDSLKK 60
61 LRLKTAQQSQ GENGSEDNER FNSKEYGFLS TKSPNEHIFS RLDLVKYDCP SLAQRIQYML 120
121 QQLEFKLQVE KQYQEANTKL TKLYQIDGDQ RSSSAAEGGA MESKYRIQML NKALKKYQAI 180
181 NVDFDQFKHQ PNDIMDNQQP KFRRKQLTGV LTIGITAARD VDHIQSPMFA RKPESYVTIK 240
241 IDDTIKARTK PSRNDRWSED FQIPVEKGNE IEITVYDKVN DSLIPVAIMW LLLSDIAEEI 300
301 RKKKAGQTNE QQGWVNASNI NGGSSLASEE GSTLTSTYSN SAIQSTSAKN VQGENTSTSQ 360
361 ISTNSWFVLE PSGQILLTLG FHKSSQIERK QLMGGLHRHG AIINRKEEIF EQHGHHFVQK 420
421 SFYNIMCCAY CGDFLRYTGF QCQDCKFLCH KKCYTNVVTK CIAKTSTDTD PDEAKLNHRI 480
481 PHRFLPTSNR GTKWCCHCGY ILPWGRHKVR KCSECGIMCH AQCAHLVPDF CGMSMEMANK 540
541 ILKTIQDTKR NQEKKKRTVP SAQLGSSIGT ANGSDLSPSK LAERANAPLP PQPRKHDKTP 600
601 SPQKVGRDSP TKQHDPIIDK KISLQTHGRE KLNKFIDENE AYLNFTEGAQ QTAEFSSPEK 660
661 TLDPTSNRRS LGLTDLSIEH SQTWESKDDL MRDELELWKA QREEMELEIK QDSGEIQEDL 720
721 EVDHIDLETK QKLDWENKND FREADLTIDS THTNPFRDMN SETFQIEQDH ASKEVLQETV 780
781 SLAPTSTHPS RTTDQQSPQK SQTSTSAKHK KRAAKRRKVS LDNFVLLKVL GKGNFGKVIL 840
841 SKSKNTDRLC AIKVLKKDNI IQNHDIESAR AEKKVFLLAT KTKHPFLTNL YCSFQTENRI 900
901 YFAMEFIGGG DLMWHVQNQR LSVRRAKFYA AEVLLALKYF HDNGVIYRDL KLENILLTPE 960
961 GHIKIADYGL CKDEMWYGNR TSTFCGTPEF MAPEILKEQE YTKAVDWWAF GVLLYQMLLC1020
1021 QSPFSGDDED EVFNAILTDE PLYPIDMAGE IVQIFQGLLT KDPEKRLGAG PRDADEVMEE1080
1081 PFFRNINFDD ILNLRVKPPY IPEIKSPEDT SYFEQEFTSA PPTLTPLPSV LTTSQQEEFR1140
1141 GFSFMPDDLD L |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
FOL3 PKC1 SSL2 TYR1 |
| View Details | Gavin AC, et al. (2002) |

|
ENO1 PKC1 |
| View Details | Qiu et al. (2008) |

|
AKL1 PKC1 SSK2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #34 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
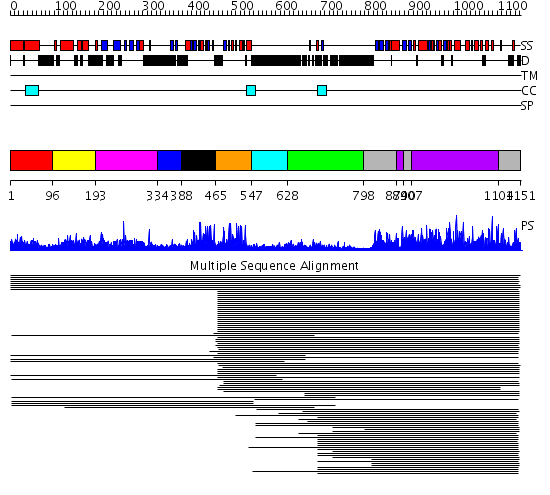
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..95] | 21.0 | Hr1 repeat No confident structure predictions are available. | |
| 2 | View Details | [96..192] | 9.8 | Effector domain of the protein kinase pkn/prk1 | |
| 3 | View Details | [193..333] | 20.221849 | Domain from protein kinase C epsilon | |
| 4 | View Details | [334..387] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [388..464] | 17.30103 | Phorbol esters/diacylglycerol binding domain (C1 domain) No confident structure predictions are available. | |
| 6 | View Details | [465..546] | 47.522879 | Protein kinase c-gamma | |
| 7 | View Details | [547..627] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [628..797] | 12.154902 | C2 domain from protein kinase c (alpha) | |
| 9 | View Details | [798..873] [890..906] [1104..1151] |
690.228787 | cAMP-dependent PK, catalytic subunit | |
| 10 | View Details | [874..889] [907..1103] |
690.228787 | cAMP-dependent PK, catalytic subunit |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)