













| General Information: |
|
| Name(s) found: |
DLDH2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 507 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
mitochondrial respiratory chain complex I [IDA] mitochondrial matrix [ISS] mitochondrion [IDA] |
| Biological Process: |
response to cadmium ion
[IEP]
|
| Molecular Function: |
ATP binding
[IDA]
zinc ion binding [IDA] copper ion binding [IDA] dihydrolipoyl dehydrogenase activity [IMP][ISS] cobalt ion binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAMASLARRK AYFLTRNISN SPTDAFRFSF SLTRGFASSG SDDNDVVIIG GGPGGYVAAI 60
61 KAAQLGLKTT CIEKRGALGG TCLNVGCIPS KALLHSSHMY HEAKHVFANH GVKVSSVEVD 120
121 LPAMLAQKDT AVKNLTRGVE GLFKKNKVNY VKGYGKFLSP SEVSVDTIDG ENVVVKGKHI 180
181 IVATGSDVKS LPGITIDEKK IVSSTGALSL TEIPKKLIVI GAGYIGLEMG SVWGRLGSEV 240
241 TVVEFAADIV PAMDGEIRKQ FQRSLEKQKM KFMLKTKVVG VDSSGDGVKL IVEPAEGGEQ 300
301 TTLEADVVLV SAGRTPFTSG LDLEKIGVET DKGGRILVNE RFSTNVSGVY AIGDVIPGPM 360
361 LAHKAEEDGV ACVEFIAGKH GHVDYDKVPG VVYTYPEVAS VGKTEEQLKK EGVSYNVGKF 420
421 PFMANSRAKA IDTAEGMVKI LADKETDKIL GVHIMSPNAG ELIHEAVLAI NYDASSEDIA 480
481 RVCHAHPTMS EAIKEAAMAT YDKPIHM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
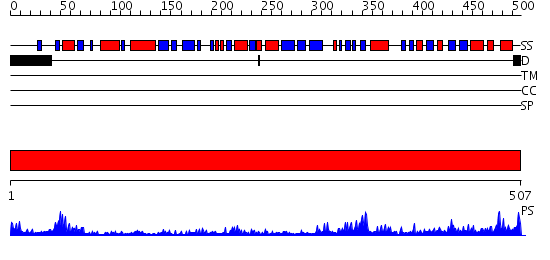
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..507] | 115.0 | Dihydrolipoamide dehydrogenase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)