













| General Information: |
|
| Name(s) found: |
CSN10_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 645 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDEDNNYDD FMLSDDEGME SIEMEEETDD EDKQNIEINE DNSQDDQDRG AARHKQHEQG 60
61 TFEKHDRVED ICERIFEQGQ ALKEDERYKE ARDLFLKIYY KEEFSSDESI ERLMTWKFKS 120
121 LIEILRLRAL QLYFQKNGAQ DLVLQILEDT ATMSVFLQRI DFQIDGNIFE LLSDTFEVLA 180
181 PKWERVFLFD IEKVDRENMI CKIDFQKNFM DQFQWILRKP GKDCKLQNLQ RIIRKKIFIA 240
241 VVWYQRLTMG NVFTPEISSQ IEILVKDNEC SSFEENNDLE SVSMLLQYYI LEYMNTARIN 300
301 NRRLFKKCID FFEMLISKSL TFSQESGLMV ILYTSKIVFI LDSDSENDLS FALMRYYDRK 360
361 EELKNMFLYI LKHLEEMGKL RERDITSLFH KFILSGFIFT SMILEAISTD KINPFGFEQV 420
421 KIALGSPIVN VLEDVYRCFA QLELRQLNAS ISLIPELSVV LSGIIQDIYY LAQTLKLWRK 480
481 IARLYSCISI SDIISMLQIS DDNEMTRDDL LTILMRSIMK NRSVVYFKLD LTSDLVYFGD 540
541 ENKVMLPRCS KEEFRLMISP KDEETTEKAR LIDFEYVNDV AIYNNPTRIR TKSSKEFFNT 600
601 LRKSRETVKL PRVSNQSNED TFLPSYMKFS NKYLELCKLA SNNLE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
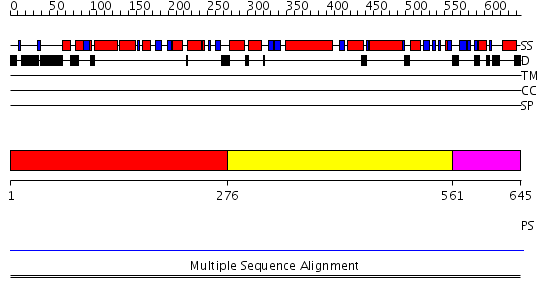
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..275] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [276..560] | 8.19 | Hemolysin E (HlyE, ClyA, SheA) | |
| 3 | View Details | [561..645] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [414..559] | 1.38 | Solution structure of the PCI domain | |
| 5 | View Details | [560..645] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.69 |
Source: Reynolds et al. (2008)