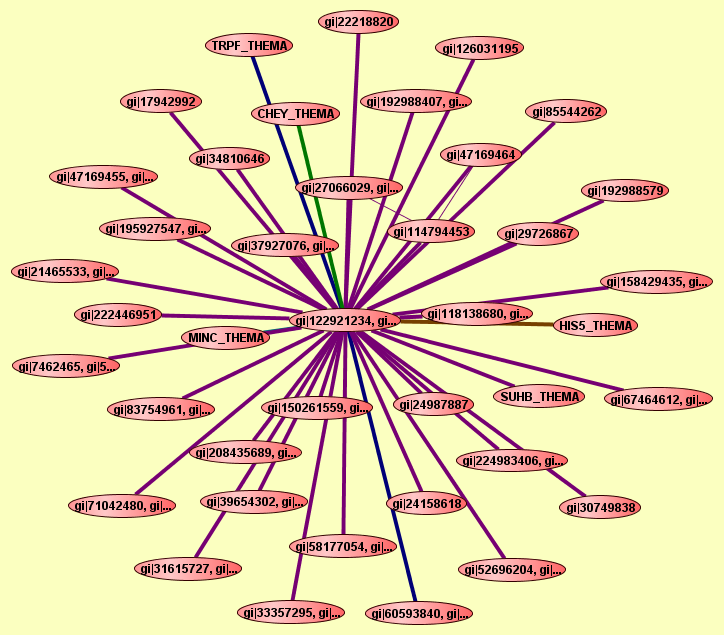
| |
Size |
Notes |
Members |
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1NSJA and 2NZCA with confidence: 0.919565061116 |
TRPF_THEMA
gi|122921233, gi...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1J5UA and 2NZCA with confidence: 0.905716052828 |
gi|122921233, gi...
gi|22218820
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 1GXJA with confidence: 0.940974017673 |
gi|122921233, gi...
gi|21465533, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 2PIBA with confidence: 0.950434841547 |
gi|122921233, gi...
gi|158429435, gi...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 2HP7A with confidence: 0.996062843372 |
gi|114794453
gi|122921233, gi...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 1VJRA with confidence: 0.959228471164 |
gi|122921233, gi...
gi|47169464
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 1K9VF with confidence: 0.92850195981 |
HIS5_THEMA
gi|122921233, gi...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 2NRRA with confidence: 0.946633672916 |
gi|122921233, gi...
gi|126031195
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 2Q78A with confidence: 0.903493562641 |
gi|122921233, gi...
gi|150261559, gi...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 2W6RA with confidence: 0.905776727686 |
gi|122921233, gi...
gi|222446951
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 1O3UA with confidence: 0.94034405021 |
gi|122921233, gi...
gi|30749838
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 1VJLA with confidence: 0.923023321884 |
gi|122921233, gi...
gi|47169456, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 1O0WA with confidence: 0.991660931515 |
gi|122921233, gi...
gi|27066029, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 3BEXA with confidence: 0.929457573397 |
gi|122921233, gi...
gi|192988420, gi...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 2ETHA with confidence: 0.913965643882 |
gi|122921233, gi...
gi|83754962, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 1HF2A with confidence: 0.914211365023 |
MINC_THEMA
gi|122921233, gi...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 1ZTCA with confidence: 0.92801804335 |
gi|122921233, gi...
gi|71042483, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 3DCZA with confidence: 0.965224571285 |
gi|122921233, gi...
gi|192988579
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 1LMEA with confidence: 0.956957335161 |
gi|122921233, gi...
gi|33357296, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 2ZVRA with confidence: 0.966829079826 |
gi|122921233, gi...
gi|224983406, gi...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 1KGSA with confidence: 0.934510736457 |
gi|122921233, gi...
gi|17942992
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 1NV8A with confidence: 0.920947470132 |
gi|122921233, gi...
gi|31615728, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 3DCLA with confidence: 0.926848514695 |
gi|122921233, gi...
gi|195927548, gi...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 1TMYA with confidence: 0.942148841296 |
CHEY_THEMA
gi|122921233, gi...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 1VLHA with confidence: 0.945579156089 |
gi|122921233, gi...
gi|52696203, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 1GUIA with confidence: 0.921816210223 |
gi|122921233, gi...
gi|24158618
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 1O1YA with confidence: 0.912530512099 |
gi|122921233, gi...
gi|29726867
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 1O5KA with confidence: 0.924230809517 |
gi|122921233, gi...
gi|37927075, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 1VQ0A with confidence: 0.949397772944 |
gi|122921233, gi...
gi|58177053, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 2P3NA with confidence: 0.914728205978 |
SUHB_THEMA
gi|122921233, gi...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 1S12A with confidence: 0.948592371898 |
gi|122921233, gi...
gi|58176698, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 2J73A with confidence: 0.91423961534 |
gi|118138678, gi...
gi|122921233, gi...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 2BBHA with confidence: 0.923369124764 |
gi|122921233, gi...
gi|85544262
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 3E57A with confidence: 0.928016784721 |
gi|122921233, gi...
gi|208435688, gi...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 1O63A with confidence: 0.922366210314 |
gi|122921233, gi...
gi|39654303, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 1WA3A with confidence: 0.915069236458 |
gi|122921233, gi...
gi|60593842, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 1ZKOA with confidence: 0.935596786262 |
gi|122921233, gi...
gi|67464612, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 1O0XA with confidence: 0.944606747142 |
gi|122921233, gi...
gi|24987887
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2NZCA and 1O50A with confidence: 0.940434083399 |
gi|122921233, gi...
gi|34810646
|