













| General Information: |
|
| Name(s) found: |
Vha100-1-PH /
FBpp0084747
[FlyBase]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 825 amino acids |
Gene Ontology: |
|
| Cellular Component: |
vacuolar proton-transporting V-type ATPase, V0 domain
[ISS]
synapse [IDA] |
| Biological Process: |
ATP synthesis coupled proton transport
[IEA]
|
| Molecular Function: |
calmodulin binding
[IDA]
hydrogen-exporting ATPase activity, phosphorylative mechanism [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTCHGHTRFI VRKFVGCWSL WTNLYLIFKH LLNPDVNAFQ RKFVNEVRRC DEMERKLRYL 60
61 EKEIKKDGIP MLDTGESPEA PQPREMIDLE ATFEKLENEL REVNQNAEAL KRNFLELTEL 120
121 KHILRKTQVF FDEMADNQNE DEQAQLLGEE GVRASQPGQN LKLGFVAGVI LRERLPAFER 180
181 MLWRACRGNV FLRQAMIETP LEDPTNGDQV HKSVFIIFFQ GDQLKTRVKK ICEGFRATLY 240
241 PCPEAPADRR EMAMGVMTRI EDLNTVLGQT QDHRHRVLVA AAKNLKNWFV KVRKIKAIYH 300
301 TLNLFNLDVT QKCLIAECWV PLLDIETIQL ALRRGTERSG SSVPPILNRM QTFENPPTYN 360
361 RTNKFTKAFQ ALIDAYGVAS YREMNPAPYT IITFPFLFAV MFGDLGHGAI MALFGLWMIR 420
421 KEKGLAAQKT DNEIWNIFFG GRYIIFLMGV FSMYTGLIYN DIFSKSLNIF GSHWHLSYNK 480
481 STVMENKFLQ LSPKGDYEGA PYPFGMDPIW QVAGANKIIF HNAYKMKISI IFGVIHMIFG 540
541 VVMSWHNHTY FRNRISLLYE FIPQLVFLLL LFFYMVLLMF IKWIKFAATN DKPYSEACAP 600
601 SILITFIDMV LFNTPKPPPE NCETYMFMGQ HFIQVLFVLV AVGCIPVMLL AKPLLIMQAR 660
661 KQANVQPIAG ATSDAEAGGV SNSGSHGGGG GHEEEEELSE IFIHQSIHTI EYVLGSVSHT 720
721 ASYLRLWALS LAHAQLAEVL WTMVLSIGLK QEGPVGGIVL TCVFAFWAIL TVGILVLMEG 780
781 LSAFLHTLRL HWVEFQSKFY KGQGYAFQPF SFDAIIENGA AAAEE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
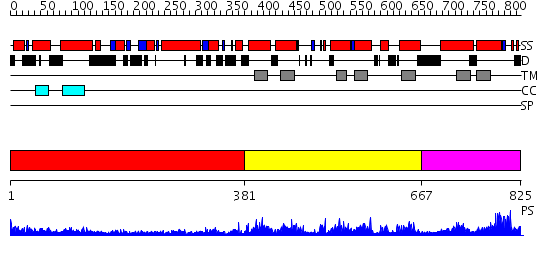
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..380] | 29.154902 | Heavy meromyosin subfragment | |
| 2 | View Details | [381..666] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [667..825] | 2.108949 | View MSA. No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|