













| General Information: |
|
| Name(s) found: |
FBpp0300539
[FlyBase]
sdt-PD / FBpp0088913 [FlyBase] |
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 879 amino acids |
Gene Ontology: |
|
| Cellular Component: |
apicolateral plasma membrane
[IDA]
adherens junction [IDA] subapical complex [TAS] cell-cell adherens junction [NAS] cytoplasm [NAS] membrane [ISS][NAS] septate junction [TAS] apical cortex [IDA] plasma membrane [ISS] apical plasma membrane [IDA][NAS][TAS] |
| Biological Process: |
[NOT]establishment or maintenance of neuroblast polarity
[IEP][IMP]
cell-cell junction assembly [NAS] morphogenesis of an epithelium [NAS][TAS] zonula adherens assembly [IMP][TAS] establishment or maintenance of epithelial cell apical/basal polarity [IEP][IMP][IPI][TAS] establishment or maintenance of cell polarity [NAS] morphogenesis of a polarized epithelium [TAS] establishment or maintenance of polarity of embryonic epithelium [IMP] |
| Molecular Function: |
protein binding
[IPI][ISS][TAS]
guanylate kinase activity [ISS][NAS][TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVVSTLHITL ENNGNVAAGQ PAAVAAASVV GAATSPTKFA VLSAQQLQLQ LQQQQQQQNE 60
61 KENHMNNNNN NSDYCDMNGN GVGMGNGGSG GPGSLTPQSP DQYVGSPVSS AATPQRTASA 120
121 LRRGIQIQRS STRLRQQQEQ LRRRREEEER IAQQNEFLRN SLRGSRKLKA LQDTATPGKA 180
181 VAQQQQQATL ATQVVGVENE AYLPDEDQPQ AEQIDGYGEL IAALTRLQNQ LSKSGLSTLA 240
241 GRVSAAHSVL ASASVAHVLA ARTAVLQRRR SRVSGPLHHS SLGLQKDIVE LLTQSNTAAA 300
301 IELGNLLTSH EMEGLLLAHD RIANHTDGTP SPTPTPTPAI GAATGSTLSS PVAGPKRNLG 360
361 MVVPPPVVPP PLAQRGAMPL PRGESPPPVP MPPLATMPMS MPVNLPMSAC FGTLNDQNDN 420
421 IRIIQIEKST EPLGATVRNE GEAVVIGRIV RGGAAEKSGL LHEGDEILEV NGQELRGKTV 480
481 NEVCALLGAM QGTLTFLIVP AGSPPSVGVM GGTTGSQLAG LGGAHRDTAV LHVRAHFDYD 540
541 PEDDLYIPCR ELGISFQKGD VLHVISREDP NWWQAYREGE EDQTLAGLIP SQSFQHQRET 600
601 MKLAIAEEAG LARSRGKDGS GSKGATLLCA RKGRKKKKKA SSEAGYPLYA TTAPDETDPE 660
661 EILTYEEVAL YYPRATHKRP IVLIGPPNIG RHELRQRLMA DSERFSAAVP HTSRARREGE 720
721 VPGVDYHFIT RQAFEADILA RRFVEHGEYE KAYYGTSLEA IRTVVASGKI CVLNLHPQSL 780
781 KLLRASDLKP YVVLVAPPSL DKLRQKKLRN GEPFKEEELK DIIATARDME ARWGHLFDMI 840
841 IINNDTERAY HQLLAEINSL EREPQWVPAQ WVHNNRDES |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
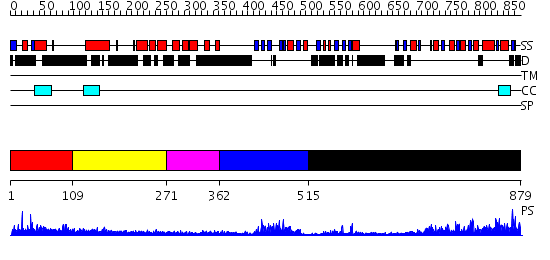
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..108] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [109..270] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [271..361] | 1.28 | NMR structure of L27 heterodimer from C. elegans Lin-7 and H. sapiens Lin-2 scaffold proteins | |
| 4 | View Details | [362..514] | 15.045757 | Solution structure of the PDZ domain of Pals1 protein | |
| 5 | View Details | [515..879] | 51.0 | Psd-95; Guanylate kinase-like domain of Psd-95 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.54 |
Source: Reynolds et al. (2008)