













| General Information: |
|
| Name(s) found: |
sdt-PE /
FBpp0088912
[FlyBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1367 amino acids |
Gene Ontology: |
|
| Cellular Component: |
apicolateral plasma membrane
[IDA]
adherens junction [IDA] subapical complex [TAS] cell-cell adherens junction [NAS] cytoplasm [NAS] membrane [ISS][NAS] septate junction [TAS] apical cortex [IDA] plasma membrane [ISS] apical plasma membrane [IDA][NAS][TAS] |
| Biological Process: |
[NOT]establishment or maintenance of neuroblast polarity
[IEP][IMP]
cell-cell junction assembly [NAS] morphogenesis of an epithelium [NAS][TAS] zonula adherens assembly [IMP][TAS] establishment or maintenance of epithelial cell apical/basal polarity [IEP][IMP][IPI][TAS] establishment or maintenance of cell polarity [NAS] morphogenesis of a polarized epithelium [TAS] establishment or maintenance of polarity of embryonic epithelium [IMP] |
| Molecular Function: |
protein binding
[IPI][ISS][TAS]
guanylate kinase activity [ISS][NAS][TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRILKQWNRR RSGSSIVVLD GDDLKPCLPD DYISGQHHLN HQQQLQLQQQ LQQQHPLQQQ 60
61 HYRTHSGDIR EIDQEMLTML SVNQDNGPHR EMAVDCPDTF IARNKTPPRY PPPRPPQKHK 120
121 KSTNTTTTTT ITALTNNDHA NKMLIVAYHS SHQHEQLQQQ HPSKTSTTTT TIALDVATQN 180
181 LYNQKQQNKL EQIENYENCL QSERNEQHEQ QFEQQKQHQA TTAMAATQVA QQQTPSHKLQ 240
241 ATLSSDPNGN SNSNNNSHIV GISSSSSSNN SSITDDFLCV VDGLYQGRKD TASPSSSAFD 300
301 EVMSKHTLDS FGSIAYRHLH QQHQATSNGN SSSNTSNTNS NTNSNTNSNS NTNGNTSNNT 360
361 AVSTKTATVT KTGVSSSNSN SNSLNSSNSS MHTSSSSSGH SSNIASATSS SSATSSSTVP 420
421 DDLSLAPPGY EVSQQQQQQH LVATPVTMLL PPMAKHRELP VDVPDSFIEM VKTTPRYPPP 480
481 AHLSSRGSLL SNGSASTAHT TLSSMGVAPS PVTATAAAAA SASAACATTA VAAAAVSGVA 540
541 DGDARRVADE LNGNAKPVPP PRDHLRVEKD GRLVNCSPAP QLPDRRAPGN ASSGSSGATT 600
601 HPLQHQQIAQ IVEPTLEQLD SIKKYQEQLR RRREEEERIA QQNEFLRNSL RGSRKLKALQ 660
661 DTATPGKAVA QQQQQATLAT QVVGVENEAY LPDEDQPQAE QIDGYGELIA ALTRLQNQLS 720
721 KSGLSTLAGR VSAAHSVLAS ASVAHVLAAR TAVLQRRRSR VSGPLHHSSL GLQKDIVELL 780
781 TQSNTAAAIE LGNLLTSHEM EGLLLAHDRI ANHTDGTPSP TPTPTPAIGA ATGSTLSSPV 840
841 AGPKRNLGMV VPPPVVPPPL AQRGAMPLPR GESPPPVPMP PLATMPMSMP VNLPMSACFG 900
901 TLNDQNDNIR IIQIEKSTEP LGATVRNEGE AVVIGRIVRG GAAEKSGLLH EGDEILEVNG 960
961 QELRGKTVNE VCALLGAMQG TLTFLIVPAG SPPSVGVMGG TTGSQLAGLG GAHRDTAVLH1020
1021 VRAHFDYDPE DDLYIPCREL GISFQKGDVL HVISREDPNW WQAYREGEED QTLAGLIPSQ1080
1081 SFQHQRETMK LAIAEEAGLA RSRGKDGSGS KGATLLCARK GRKKKKKASS EAGYPLYATT1140
1141 APDETDPEEI LTYEEVALYY PRATHKRPIV LIGPPNIGRH ELRQRLMADS ERFSAAVPHT1200
1201 SRARREGEVP GVDYHFITRQ AFEADILARR FVEHGEYEKA YYGTSLEAIR TVVASGKICV1260
1261 LNLHPQSLKL LRASDLKPYV VLVAPPSLDK LRQKKLRNGE PFKEEELKDI IATARDMEAR1320
1321 WGHLFDMIII NNDTERAYHQ LLAEINSLER EPQWVPAQWV HNNRDES |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
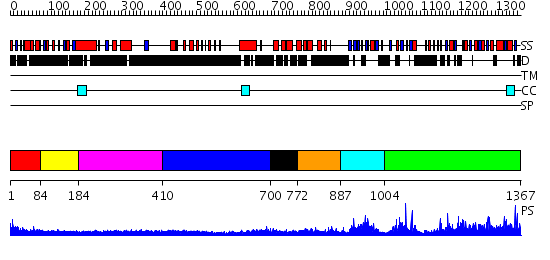
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..83] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [84..183] | 2.193997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [184..409] | 2.197996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [410..699] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [700..771] | 0.965 | 2.1 Angstrom crystal structure of the PALS-1-L27N and PATJ L27 heterodimer complex | |
| 6 | View Details | [772..886] | 6.39794 | NMR structure of L27 heterodimer from C. elegans Lin-7 and H. sapiens Lin-2 scaffold proteins | |
| 7 | View Details | [887..1003] | 20.30103 | Solution structure of the PDZ domain of Pals1 protein | |
| 8 | View Details | [1004..1367] | 93.0 | Psd-95; Guanylate kinase-like domain of Psd-95 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 7 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)