













| General Information: |
|
| Name(s) found: |
gi|55961106
[NCBI NR]
gi|42544179 [NCBI NR] gi|196115147 [NCBI NR] gi|133923367 [NCBI NR] gi|119608164 [NCBI NR] gi|119608159 [NCBI NR] |
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 898 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFSQQQQQQL QQQQQQLQQL QQQQLQQQQL QQQQLLQLQQ LLQQSPPQAP LPMAVSRGLP 60
61 PQQPQQPLLN LQGTNSASLL NGSMLQRALL LQQLQGLDQF AMPPATYDTA GLTMPTATLG 120
121 NLRGYGMASP GLAAPSLTPP QLATPNLQQF FPQATRQSLL GPPPVGVPMN PSQFNLSGRN 180
181 PQKQARTSSS TTPNRKDSSS QTMPVEDKSD PPEGSEEAAE PRMDTPEDQD LPPCPEDIAK 240
241 EKRTPAPEPE PCEASELPAK RLRSSEEPTE KEPPGQLQVK AQPQARMTVP KQTQTPDLLP 300
301 EALEAQVLPR FQPRVLQVQA QVQSQTQPRI PSTDTQVQPK LQKQAQTQTS PEHLVLQQKQ 360
361 VQPQLQQEAE PQKQVQPQVQ PQAHSQGPRQ VQLQQEAEPL KQVQPQVQPQ AHSQPPRQVQ 420
421 LQLQKQVQTQ TYPQVHTQAQ PSVQPQEHPP AQVSVQPPEQ THEQPHTQPQ VSLLAPEQTP 480
481 VVVHVCGLEM PPDAVEAGGG MEKTLPEPVG TQVSMEEIQN ESACGLDVGE CENRAREMPG 540
541 VWGAGGSLKV TILQSSDSRA FSTVPLTPVP RPSDSVSSTP AATSTPSKQA LQFFCYICKA 600
601 SCSSQQEFQD HMSEPQHQQR LGEIQHMSQA CLLSLLPVPR DVLETEDEEP PPRRWCNTCQ 660
661 LYYMGDLIQH RRTQDHKIAK QSLRPFCTVC NRYFKTPRKF VEHVKSQGHK DKAKELKSLE 720
721 KEIAGQDEDH FITVDAVGCF EGDEEEEEDD EDEEEIEVEE ELCKQVRSRD ISREEWKGSE 780
781 TYSPNTAYGV DFLVPVMGYI CRICHKFYHS NSGAQLSHCK SLGHFENLQK YKAAKNPSPT 840
841 TRPVSRRCAI NARNALTALF TSSGRPPSQP NTQDKTPSKV TARPSQPPLP RRSTRLKT |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
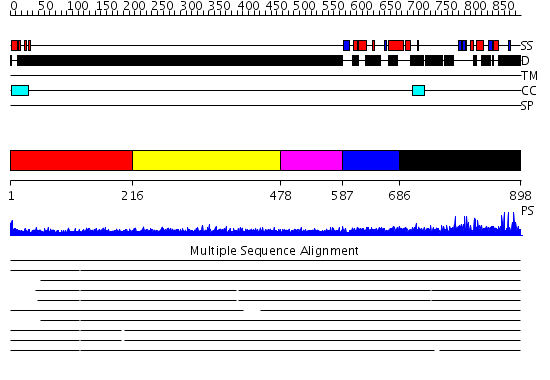
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..215] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [216..477] | 3.257996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [478..586] | 4.245996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [587..685] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [686..898] | 2.07 | Solution Structure of the N-terminal Zinc Fingers of the Xenopus laevis double stranded RNA binding protein ZFa |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)