













| General Information: |
|
| Name(s) found: |
YNBC_ECOLI
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Escherichia coli |
| Length: | 585 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MENSRIPGEH FFTTSDNTAL FYRHWPALQP GAKKVIVLFH RGHEHSGRLQ HLVDELAMPD 60
61 TAFYAWDARG HGKSSGPRGY SPSLARSVRD VDEFVRFAAS DSQVGLEEVV VIAQSVGAVL 120
121 VATWIHDYAP AIRGLVLASP AFKVKLYVPL ARPALALWHR LRGLFFINSY VKGRYLTHDR 180
181 QRGASFNNDP LITRAIAVNI LLDLYKTSER IIRDAAAITL PTQLLISGDD YVVHRQPQID 240
241 FYQRLRSPLK ELHLLPGFYH DTLGEENRAL AFEKMQSFIS RLYANKSQKF DYQHEDCTGP 300
301 SADRWRLLSG GPVPLSPVDL AYRFMRKAMK LFGTHSSGLH LGMSTGFDSG SSLDYVYQNQ 360
361 PQGSNAFGRL VDKIYLNSVG WRGIRQRKTH LQILIKQAVA DLHAKGLAVR VVDIAAGHGR 420
421 YVLDALANEP AVSDILLRDY SELNVAQGQE MIAQRGMSGR VRFEQGDAFN PEELSALTPR 480
481 PTLAIVSGLY ELFPENEQVK NSLAGLANAI EPGGILIYTG QPWHPQLEMI AGVLTSHKDG 540
541 KPWVMRVRSQ GEMDSLVRDA GFDKCTQRID EWGIFTVSMA VRRDN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
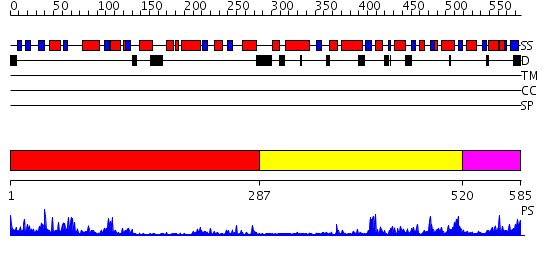
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..286] | 38.522879 | CHLOROPEROXIDASE F/PROPIONATE COMPLEX | |
| 2 | View Details | [287..519] | 14.154902 | The crystal structure of E. coli Fmu binary complex with S-Adenosylmethionine at 2.1 A resolution | |
| 3 | View Details | [520..585] | 13.09691 | Crystal Structure of Human methyltransferase AD-003 in complex with S-adenosyl-L-homocysteine |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 |
|
||||||||||||||||||
| 2 |
|
||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)