













| General Information: |
|
| Name(s) found: |
YCIR_ECOLI
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli |
| Length: | 661 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKTVRESTTL YNFLGSHNPY WRLTESSDVL RFSTTETTEP DRTLQLSAEQ AARIREMTVI 60
61 TSSLMMSLTV DESDLSVHLV GRKINKREWA GNASAWHDTP AVARDLSHGL SFAEQVVSEA 120
121 HSAIVILDSR GNIQRFNRLC EDYTGLKEHD VIGQSVFKLF MSRREAAASR RNNRVFFRSG 180
181 NAYEVELWIP TCKGQRLFLF RNKFVHSGSG KNEIFLICSG TDITEERRAQ ERLRILANTD 240
241 SITGLPNRNA MQDLIDHAIN HADNNKVGVV YLDLDNFKKV NDAYGHLFGD QLLRDVSLAI 300
301 LSCLEHDQVL ARPGGDEFLV LASNTSQSAL EAMASRILTR LRLPFRIGLI EVYTSCSVGI 360
361 ALSPEHGSDS TAIIRHADTA MYTAKEGGRG QFCVFTPEMN QRVFEYLWLD TNLRKALEND 420
421 QLVIHYQPKI TWRGEVRSLE ALVRWQSPER GLIPPLDFIS YAEESGLIVP LGRWVILDVV 480
481 RQVAKWRDKG INLRVAVNIS ARQLADQTIF TALKQVLQEL NFEYCPIDVE LTESCLIEND 540
541 ELALSVIQQF SQLGAQVHLD DFGTGYSSLS QLARFPIDAI KLDQVFVRDI HKQPVSQSLV 600
601 RAIVAVAQAL NLQVIAEGVE SAKEDAFLTK NGINERQGFL FAKPMPAVAF ERWYKRYLKR 660
661 A |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
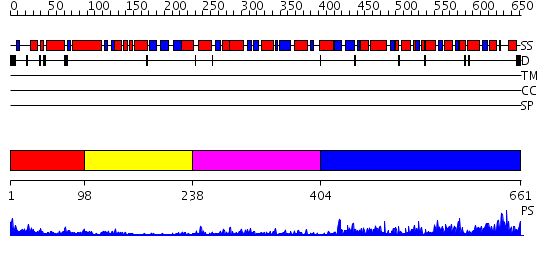
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..97] | 8.0 | No description for 2gj3A was found. | |
| 2 | View Details | [98..237] | 5.92 | Crystal Structure of the heme PAS sensor domain of Ec DOS (ferric form) | |
| 3 | View Details | [238..403] | 15.0 | Response regulator PleD in complex with c-diGMP | |
| 4 | View Details | [404..661] | 69.0 | No description for 2r6oA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)