













| General Information: |
|
| Name(s) found: |
MLRA_ECOLI
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Escherichia coli |
| Length: | 243 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALYTIGEVA LLCDINPVTL RAWQRRYGLL KPQRTDGGHR LFNDADIDRI REIKRWIDNG 60
61 VQVSKVKMLL SNENVDVQNG WRDQQETLLT YLQSGNLHSL RTWIKERGQD YPAQTLTTHL 120
121 FIPLRRRLQC QQPTLQALLA ILDGVLINYI AICLASARKK QGKDALVVGW NIQDTTRLWL 180
181 EGWIASQQGW RIDVLAHSLN QLRPELFEGR TLLVWCGENR TSAQQQQLTS WQEQGHDIFP 240
241 LGI |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
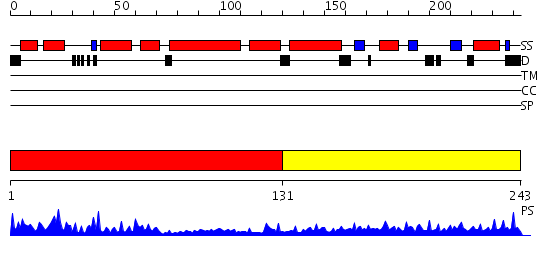
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..130] | 27.69897 | Crystal structure of the Cu(I) form of E. coli CueR, a copper efflux regulator | |
| 2 | View Details | [131..243] | 1.154987 | View MSA. Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 |
|
||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.57 |
Source: Reynolds et al. (2008)