













| General Information: |
|
| Name(s) found: |
ANP1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 666 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
response to oxidative stress
[IDA]
flower development [IGI] |
| Molecular Function: |
MAP kinase kinase kinase activity
[ISS]
kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQDFFGSVRR SLVFRPSSDD DNQENQPPFP GVLADKITSC IRKSKIFIKP SFSPPPPANT 60
61 VDMAPPISWR KGQLIGRGAF GTVYMGMNLD SGELLAVKQV LIAANFASKE KTQAHIQELE 120
121 EEVKLLKNLS HPNIVRYLGT VREDDTLNIL LEFVPGGSIS SLLEKFGPFP ESVVRTYTRQ 180
181 LLLGLEYLHN HAIMHRDIKG ANILVDNKGC IKLADFGASK QVAELATMTG AKSMKGTPYW 240
241 MAPEVILQTG HSFSADIWSV GCTVIEMVTG KAPWSQQYKE VAAIFFIGTT KSHPPIPDTL 300
301 SSDAKDFLLK CLQEVPNLRP TASELLKHPF VMGKHKESAS TDLGSVLNNL STPLPLQINN 360
361 TKSTPDSTCD DVGDMCNFGS LNYSLVDPVK SIQNKNLWQQ NDNGGDEDDM CLIDDENFLT 420
421 FDGEMSSTLE KDCHLKKSCD DISDMSIALK SKFDESPGNG EKESTMSMEC DQPSYSEDDD 480
481 ELTESKIKAF LDEKAADLKK LQTPLYEEFY NSLITFSPSC MESNLSNSKR EDTARGFLKL 540
541 PPKSRSPSRG PLGGSPSRAT DATSCSKSPG SGGSRELNIN NGGDEASQDG VSARVTDWRG 600
601 LVVDTKQELS QCVALSEIEK KWKEELDQEL ERKRQEIMRQ AGLGSSPRDR GMSRQREKSR 660
661 FASPGK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
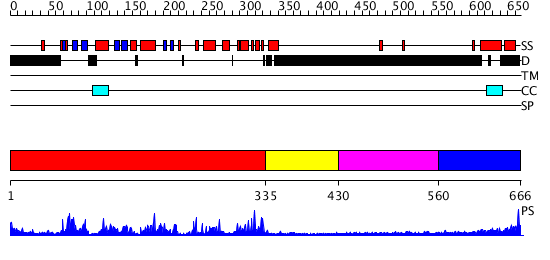
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..334] | 58.30103 | No description for 2clqA was found. | |
| 2 | View Details | [335..429] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [430..559] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [560..666] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)