













| General Information: |
|
| Name(s) found: |
RSMA_ENTFA
[Swiss-Prot]
KSGA_ENTFA [Swiss-Prot] |
| Description(s) found:
Found 6 descriptions. SHOW ALL |
|
| Organism: | Enterococcus faecalis |
| Length: | 295 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTDYKEIATP SRTKEILKKH GFSFKKSLGQ NFLTEPNILR KIVETAGINQ QTNVVEVGPG 60
61 IGALTEQLAM NAAQVVAFEI DDRLIPVLAD TLSRYDNVTV VHQDVLKADL VETTNQVFQE 120
121 KYPIKVVANL PYYITTPIMM HFLESSLDVA EMVVMMQKEV ADRIAAKPGT KAYGSLSIAV 180
181 QYFMEASVAF IVPKTVFVPQ PNVDSAIIKL TRRATPAVTV TNEKEFFKLT KASFQLRRKT 240
241 LWNNLTHFYG KDEQTVAWLK ESLAEAEIDP SRRGETLSLE EFARLSNALE KNKPV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
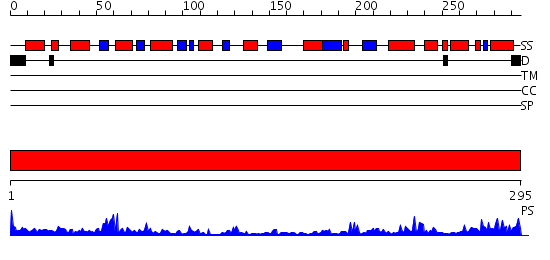
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..295] | 52.0 | 2.1 Angstrom Crystal structure of KsgA: A Universally Conserved Adenosine Dimethyltransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)