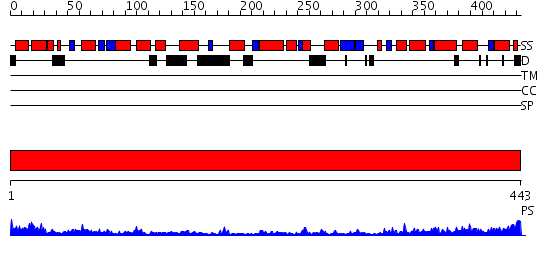
Description(s) found:
SHOW ONLY BEST
|
gi|73535733|pdb|1YYF|B Chain B, Correction Of X-Ray Intensities From An Hslv-Hslu Co- Crystal Containing Lattice Translocation Defects
[NCBI NR]
gi|73535732|pdb|1YYF|A Chain A, Correction Of X-Ray Intensities From An Hslv-Hslu Co- Crystal Containing Lattice Translocation Defects
[NCBI NR]
gi|62288339|sp|P0A6H5.1|HSLU_ECOLI RecName: Full=ATP-dependent hsl protease ATP-binding subunit hslU; AltName: Full=Heat shock protein hslU
[NCBI NR]
pir||JT0761 heat shock protein hslU - Escherichia coli (strain K-12)�
[NCBI NR]
gi|305034|gb|AAB03063.1| matches PS00017: ATP_GTP_A; similar to Pasteurella haemolytica hypoth. protein ORF1; heat shock induced [Escherichia coli]
[NCBI NR]
pir||B86083 heat shock protein HslU [imported] - Escherichia coli (strain O157:H7, substrain EDL933)�
[NCBI NR]
pir||B91236 heat shock protein HslU [imported] - Escherichia coli (strain O157:H7, substrain RIMD 0509952)�
[NCBI NR]
gi|209752256|gb|ACI74435.1| heat shock protein HslV [Escherichia coli]
[NCBI NR]
gi|209752254|gb|ACI74434.1| heat shock protein HslV [Escherichia coli]
[NCBI NR]
gi|209752252|gb|ACI74433.1| heat shock protein HslV [Escherichia coli]
[NCBI NR]
gi|209752250|gb|ACI74432.1| heat shock protein HslV [Escherichia coli]
[NCBI NR]
gi|209752248|gb|ACI74431.1| heat shock protein HslV [Escherichia coli]
[NCBI NR]
gi|13399760|pdb|1G4B|L Chain L, Crystal Structures Of The Hslvu Peptidase-Atpase Complex Reveal An Atp-Dependent Proteolysis Mechanism
[NCBI NR]
gi|13399759|pdb|1G4B|K Chain K, Crystal Structures Of The Hslvu Peptidase-Atpase Complex Reveal An Atp-Dependent Proteolysis Mechanism
[NCBI NR]
gi|13399758|pdb|1G4B|F Chain F, Crystal Structures Of The Hslvu Peptidase-Atpase Complex Reveal An Atp-Dependent Proteolysis Mechanism
[NCBI NR]
gi|13399757|pdb|1G4B|E Chain E, Crystal Structures Of The Hslvu Peptidase-Atpase Complex Reveal An Atp-Dependent Proteolysis Mechanism
[NCBI NR]
gi|13399752|pdb|1G4A|F Chain F, Crystal Structures Of The Hslvu Peptidase-Atpase Complex Reveal An Atp-Dependent Proteolysis Mechanism
[NCBI NR]
gi|13399751|pdb|1G4A|E Chain E, Crystal Structures Of The Hslvu Peptidase-Atpase Complex Reveal An Atp-Dependent Proteolysis Mechanism
[NCBI NR]
ATP-dependent hsl protease ATP-binding subunit hslU (Heat shock protein hslU) - Escherichia coli
[Swiss-Prot]
ATPase component of the HslUV protease, also functions as molecular chaperone; heat shock protein hslVU, ATPase subunit, homologous to chaperones [Escherichia coli K12]�gi|15834112|ref|NP_312885.1| HslU [Escherichia coli O157:H7]�gi|30064776|ref|NP_838947
[nrdb_11152004_Chlamydomonas]
ATP-dependent hsl protease ATP-binding subunit hslU (Heat shock protein hslU)
[NCBI-RefSeq_human_na_08-09-2006_reversed.fasta]
matches PS00017: ATP_GTP_A; similar to Pasteurella haemolytica hypoth. protein ORF1; heat shock induced [Escherichia coli]
[NCBI-RefSeq_human_na_08-09-2006_reversed.fasta]
Chain F, Crystal Structures Of The Hslvu Peptidase-Atpase Complex Reveal An Atp-Dependent Proteolysis Mechanism
[nr-271106-contam.fasta]
Chain L, Crystal Structures Of The Hslvu Peptidase-Atpase Complex Reveal An Atp-Dependent Proteolysis Mechanism
[nr-271106-contam.fasta]
Chain E, Crystal Structures Of The Hslvu Peptidase-Atpase Complex Reveal An Atp-Dependent Proteolysis Mechanism
[nr-271106-contam.fasta]
Chain A, Correction Of X-Ray Intensities From An Hslv-Hslu Co- Crystal Containing Lattice Translocation Defects
[nr-271106-contam.fasta]
Chain K, Crystal Structures Of The Hslvu Peptidase-Atpase Complex Reveal An Atp-Dependent Proteolysis Mechanism
[nr-271106-contam.fasta]
Chain B, Correction Of X-Ray Intensities From An Hslv-Hslu Co- Crystal Containing Lattice Translocation Defects
[nr-271106-contam.fasta]
ATP-dependent protease ATPase subunit HslU OS=Escherichia coli (strain K12) GN=hslU PE=1 SV=1
[uniprot-Ecoli-reviewed-110511.fasta]
|