













| General Information: |
|
| Name(s) found: |
gi|151941570
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae YJM789 |
| Length: | 305 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSEFNITETY LRFLEEDTEM TMPIAAIEAL VTLLRIKTPE TAAEMINTIK SSTEELIKSI 60
61 PNSVSLRAGC DIFMRFVLRN LHLYGDWENC KQHLIENGQL FVSRAKKSRN KIAEIGVDFI 120
121 ADDDIILVHG YSRAVFSLLN HAANKFIRFR CVVTESRPSK QGNQLYTLLE QKGIPVTLIV 180
181 DSAVGAVIDK VDKVFVGAEG VAESGGIINL VGTYSVGVLA HNARKPFYVV TESHKFVRMF 240
241 PLSSDDLPMA GPPLDFTRRT DDLEDALRGP TIDYTAQEYI TALITDLGVL TPSAVSEELI 300
301 KMWYD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
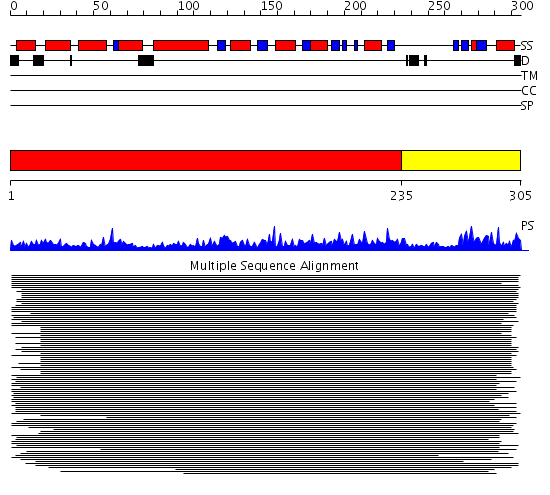
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..234] | 7.83 | Signal sequence recognition protein Ffh; Signal sequence binding protein Ffh; GTPase domain of the signal sequence recognition protein Ffh | |
| 2 | View Details | [235..305] | 10.02 | D-ribose-5-phosphate isomerase (RpiA), catalytic domain; D-ribose-5-phosphate isomerase (RpiA), lid domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)