













| General Information: |
|
| Name(s) found: |
SO0174
[shew_pnnl_plus_contams_psm.fasta]
|
| Description(s) found: |
|
| Organism: | Shewanella oneidensis |
| Length: | 395 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSERLFIRLG RTAEQACSWL VWSEQEQEII ASGELANAQG LSTLTERAGN RPIDVLVPAS 60
61 AMTLTSVNLP EKGQRQALKA LPFMLEESIA DDVDAMHFTV GPRNGDALSV VAVAHEQMQT 120
121 WLSCLADAGL KVKRIVPDCL ALPLQECQWA AMRFGNELLL RTGLGTGHSL PLPWLPVAMK 180
181 QLTAQAEELS VASYSEMQLD GVELKPQPLD LPMLVLARGI LHAPINLLSG VYTPKREYSK 240
241 HLLQWKNAAI VLAVAIVLSL VNKGLTIHQV NGQIADLKAQ SEAIYQQVVP GNTRIVNLRS 300
301 QMESQLRSLQ GQGSGAEFFS MLAGLQDAFK QVPELKPNSL RFESARNEIR MQVTAKNYAQ 360
361 IEKFKEIVGR RFQMDGGTMN SGEDQVTSTL TLRSK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
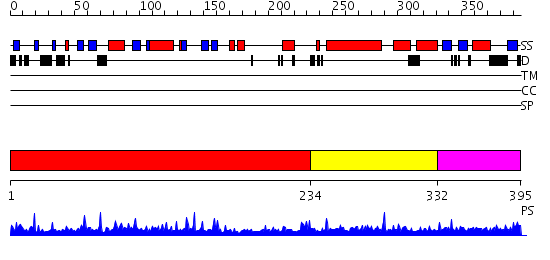
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..233] | 56.69897 | cyto-EpsL: the cytoplasmic domain of EpsL, an inner membrane component of the type II secretion system of Vibrio cholerae | |
| 2 | View Details | [234..331] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [332..395] | 1.087988 | View MSA. Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.64 |
Source: Reynolds et al. (2008)