













| General Information: |
|
| Name(s) found: |
epc-1 /
CE36670
[WormBase]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 795 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
negative regulation of vulval development
[IMP locomotory behavior [IMP nematode larval development [IMP positive regulation of growth rate [IMP embryonic development ending in birth or egg hatching [IMP gamete generation [IMP growth [IMP positive regulation of multicellular organism growth [IMP |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATTSKAFRA RALDSNRSMT VYWGHELPDL SECSVGNRAV TQMPSGMEKE EEQEKHLQEA 60
61 IAAQQASTSG IQLNHVIPTP KVDRVEDQRY HSTYHNKNKM HRSKYIKVHA WQALERDEPE 120
121 YDYDTEDEAW LSDHTHIDPR VLEKIFDTVE SHSSETQIAS EDSVINLHKS LDSSIVYEIY 180
181 EYWLSKRTSA ATTSGCVGVG GLIPRVRTEC RKDGQGVINP YVAFRRRAEK MQTRKNRKND 240
241 EDSYEKILKL VHDMSKAQQL FDMTARREKQ KLALIDMESE ILAKRMEMSD FGGSPSSFNE 300
301 ITEKIRAAAT LEVVKPPLAE INGSDEVKKR KKPRRKIADK DLISKAWLKK NAESWNRPPS 360
361 LFGQHSGNVP TVTTKPVRES LANGRFAFKR RRGCVYRAAL TVYNVPTAPA TVPPVQTQAA 420
421 VASSSSSKST DMVPSNMKFF ETFVRDSQDS VSRSLGFVRR RMGRGGRVVF DRMPRNRDDN 480
481 DERTSTDPWA EYCVADSSRT FRARNSSLGT EEETDDLSPK SLYFARSNRF AFNDDETERE 540
541 WTSRCQQSSW RDTEVDDELK KRETTSEKFT ETTTNGSTKT HTESDDSEVE RMEVDDQVDE 600
601 AQITVSSSKD DGMNGNDKNE DEEDDDDDMD VDEHQTVVGV HQHQQQQHHQ QKVRHQMNGG 660
661 GGGGGVVKLK PPLQELSPPL SGNGRADRAE PTPVPAKMCG TVSDSDDWRE PSGSPSESNS 720
721 STEWGGYTPQ EQHAVVVANA VAVAFKEKLM NGVDDDDDQQ PSPARGARDH SIKDSMSTVT 780
781 LLERKKFMMP STIGL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
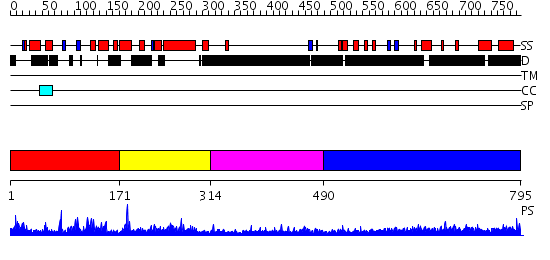
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..170] | 5.089976 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [171..313] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [314..489] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [490..795] | 1.21 | THE BLUETONGUE VIRUS (BTV) CORE BINDS DSRNA |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)