













| General Information: |
|
| Name(s) found: |
sma-9 /
CE36402
[WormBase]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 537 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA intracellular [IEA proteinaceous extracellular matrix [IEA |
| Biological Process: |
transforming growth factor beta receptor signaling pathway
[IGI tail tip morphogenesis [IMP cell adhesion [IEA blood coagulation [IEA positive regulation of multicellular organism growth [IMP |
| Molecular Function: |
nucleic acid binding
[IEA zinc ion binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLKKHIKSHT DVRAFNCTAC NFSFKTKGNL TKHLSSKTHQ RRISNIQAGN DSDGTTPSTS 60
61 SMMNMDDGYH RNQPLFDDYD NNSSDEEDYD HLNRMQAEHK FKFGQEHILF ERTAHTPPTR 120
121 WCLVEAQNDH YWPSPDRRSC MSAPPVAMQR DFDDRAMTPV SGANSPYLSQ QVHSPMSTSS 180
181 QSNIILDIPN NQKSNCSSVS NVSPSNSQNF QSLSTVPTCA SSSSNVLVPN VNFLQKDETL 240
241 KCDQCDRTFR KISDLTLHQH THNIEMQQSK NRMYQCSECK IPIRTKAQLQ KHLERNHGVH 300
301 MDESVTACID PLASTQSVLG GPSTSNPRSF MCVDCDIGFR KHGILAKHLR SKTHVMKLES 360
361 LQRLPVDTLS LITKKDNGAC LNDIDTTDCE KARISLLAIV EKLRNEADKD EQGSVVPTTS 420
421 IPAPQPVALT PEMIRALANA QTPVTASMTN TPSTAQFPVG VVSTPSQLQQ LHYVAQSNCG 480
481 STGVSNTKSV IPIGIVLNAV QKESSFGLGN TRERHAQNNS TNLGDSVQST SQSSTSS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
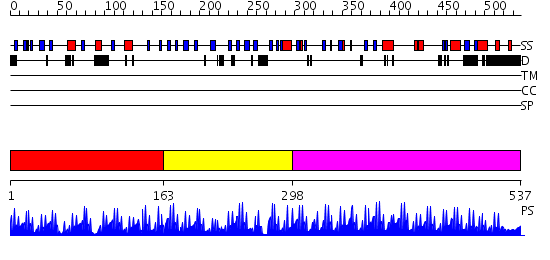
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..162] | 21.522879 | Five-finger GLI1 | |
| 2 | View Details | [163..297] | 14.154902 | No description for 2yt9A was found. | |
| 3 | View Details | [298..537] | 57.522879 | No description for 2i13A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)