













| General Information: |
|
| Name(s) found: |
sma-9 /
CE36396
[WormBase]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 725 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA intracellular [IEA proteinaceous extracellular matrix [IEA |
| Biological Process: |
transforming growth factor beta receptor signaling pathway
[IGI tail tip morphogenesis [IMP cell adhesion [IEA blood coagulation [IEA positive regulation of multicellular organism growth [IMP |
| Molecular Function: |
nucleic acid binding
[IEA zinc ion binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAFAPGSGGG GAPPGQQPAQ MMMPQNISAE TYHRLNQVQR EQLNHQRLLQ AQLQTNGPGS 60
61 VSQQQQASQQ QQQVQHVQQQ QQSQQQQQQV ASQQNQPQLQ MNAQILQALS TPQGQNLVNM 120
121 LLMQQALNAQ QTDTQNPQQI MQHQLAQQQA QQAQQAQQAQ QARQQAEQQA QAQARHQAEQ 180
181 QAQQQAQQQA QARQQEQQAQ LAAIQQQVTP QQFAQILHMQ QQLQQQQFQQ QQLQQQQLQQ 240
241 QQLQQQQLQQ QQLQQQQLQQ VLQISQAQQQ AQQAQHVQSR QMQPSQQSQV QAQLQQQQQL 300
301 QQQQAQQLSQ QQAQQQQQLQ QLQLQQFLQQ QQQLHQQRAA AQQAQAQNNA SQQRPSVAST 360
361 PALSSTPQLN DLTQTMQAQL QQQLLLQQQQ AQAQQAQQAQ QAQLAQQAQQ QQQGQSQNRT 420
421 VSQALQYIQS MQLQQRADGT PNAPSLSKPL EQPSSSKAAS SGNESMSDHI SRIISENEVI 480
481 LQGDPVIRKK RPYHRQIGAQ SSVDHDSNSG GSTRTSPGPK DSRMLQAASR SQSLFELSGS 540
541 KHFMGSLTSG QPLLRPIQAH NDPNYTPECI YCKLTFPNEA GLQAHEVVCG KKKELEKAQI 600
601 AQEGNPHSAL KRRHTHQDAT LAMHSPLAAH TPSNMPGPSE PAIKLKKDDS TELDGTSKPD 660
661 ALQSSSSFPR SLPKEWEQHM LTLQNLAAII PPFVSERLYR IHTKDKTPEI KLVSIFGSFY 720
721 FLILK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
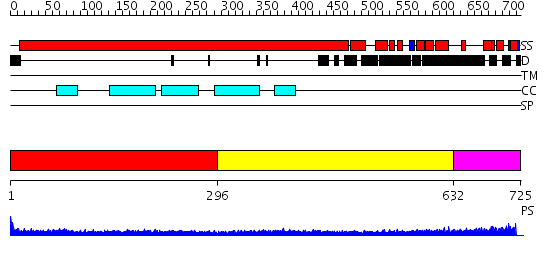
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..295] | 18.522879 | Heavy meromyosin subfragment | |
| 2 | View Details | [296..631] | 9.0 | Tropomyosin | |
| 3 | View Details | [632..725] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)