













| General Information: |
|
| Name(s) found: |
CE37018
[WormBase]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 600 amino acids |
Gene Ontology: |
|
| Cellular Component: |
transcription factor complex
[IEA |
| Biological Process: |
DNA repair
[IMP regulation of transcription, DNA-dependent [IEA |
| Molecular Function: |
transcription factor activity
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTTIFGSSNA MFLNIEANKE NIPPGTEFKK ESFVLPEPRV NRTTDPDHEN LLPSPVPRPS 60
61 PAMSQLSDAS FLSQDGGAVN TSADEADDDL EVTSRKEKSL GLLCQRFLIA INEETVGSST 120
121 REVHLETVAR KMNVEKRRIY DIVNVMEALD AMQKTNKSYY QWQGLESLPK LMFDLQNEAV 180
181 EEGLPERVLR VEQAMCSFTE LSSPRGKQGF KDIVGSFVSC TSTPTTPSTS FDSVTVKMEP 240
241 AGLLEKRSRV DTRDRQGRNS LAQLCRRFLM VLLSNPKNIR KVSLDVASTV LIKDPETEGF 300
301 EPPSRSRCRR LYDIANVLVA LGLIKKVHYL FGTKKIPLFV YCGPEPDQTG SFDVFQSVER 360
361 LLSSSQNVPQ TPIIKAQTDK IVQQIAGFGK RTLSEQNLTK PSGNTPKIAK IKSAAVRSSP 420
421 MYMENNLFMF AEVVTAEAAA EKMRYDAFAQ LSRTILGSVA SINSLNAPLT SSQHPMTSRL 480
481 PPLPMAPLQL PKLPEIPKPQ AQKQFTFPAT SRPRFNFPDY TPVQSTIRPL VSQMTFPQES 540
541 SQLHNLDVKP KHLMSNILGE SKKFKNNQNT FEHTTSSAFQ VVKKGETRPK KVFGEIQNLQ 600
601 |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
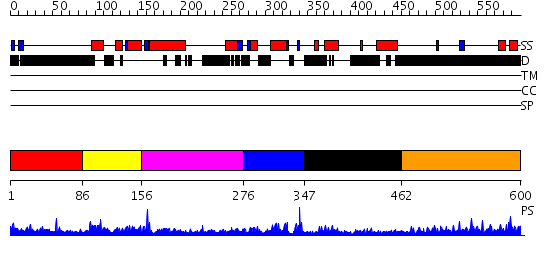
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..85] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [86..155] | 23.522879 | Cell cycle transcription factor E2F-4 | |
| 3 | View Details | [156..275] | 13.154902 | Structure of the Rb C-terminal domain bound to an E2F1-DP1 heterodimer | |
| 4 | View Details | [276..346] | 2.36 | Cell cycle transcription factor E2F-4 | |
| 5 | View Details | [347..461] | 8.154902 | Structure of the Rb C-terminal domain bound to an E2F1-DP1 heterodimer | |
| 6 | View Details | [462..600] | 5.105997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 |
|
|||||||||||||||
| 3 | No functions predicted. | |||||||||||||||
| 4 |
|
|||||||||||||||
| 5 | No functions predicted. | |||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)