













| General Information: |
|
| Name(s) found: |
AB8I_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 557 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
|
| Biological Process: |
cellular iron ion homeostasis
[IDA][TAS]
iron-sulfur cluster assembly [IGI] |
| Molecular Function: |
ATPase activity, coupled to transmembrane movement of substances
[IDA][ISS]
protein binding [IPI] transporter activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASLLANGIS SFSPQPTSDS SKSPKGFHPK PESLKFPSPK SLNPTRPIFK LRADVGIDSR 60
61 PIGASESSSS GTSTVSSTDK LQQYFQNLDY DKKYGFVEDI DSFTIPKGLS EETIRLISKL 120
121 KEEPDWMLEF RFKAYAKFLK LEEPKWSDNR YPSINFQDMC YYSAPKKKPT LNSLDEVDPQ 180
181 LLEYFDKLGV PLTEQKRLAN VAVDAVIDSV SIATTHRKTL EKSGVIFCSI SEAIREYPDL 240
241 IKKYLGRVVP SDDNYYAALN SAVFSDGSFC YIPKNTRCPM PISTYFRINA METGQFERTL 300
301 IVAEEGSFVE YLEGCTAPSY DTNQLHAAVV ELYCGKGAEI KYSTVQNWYA GDEQGKGGIY 360
361 NFVTKRGLCA GDRSKISWTQ VETGSAITWK YPSVVLEGDD SVGEFYSVAL TNNYQQADTG 420
421 TKMIHKGKNT KSRIISKGIS AGHSRNCYRG LVQVQSKAEG AKNTSTCDSM LIGDKAAANT 480
481 YPYIQVKNPS AKVEHEASTS KIGEDQLFYF QQRGIDHERA LAAMISGFCR DVFNKLPDEF 540
541 GAEVNQLMSI KLEGSVG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
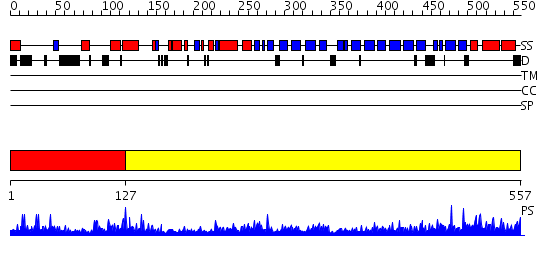
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..126] | 2.183984 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [127..557] | 75.154902 | Crystal structure of a stabilizer of iron transporter |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.69 |
Source: Reynolds et al. (2008)